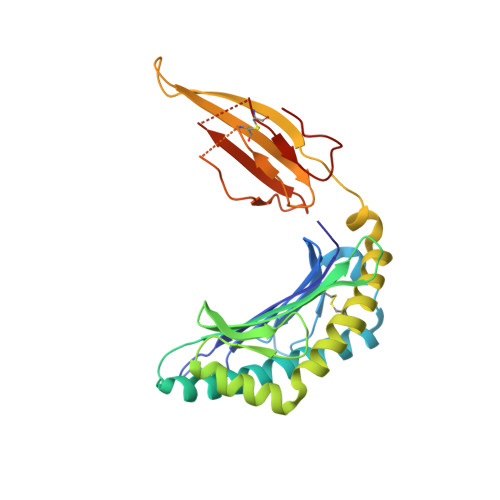
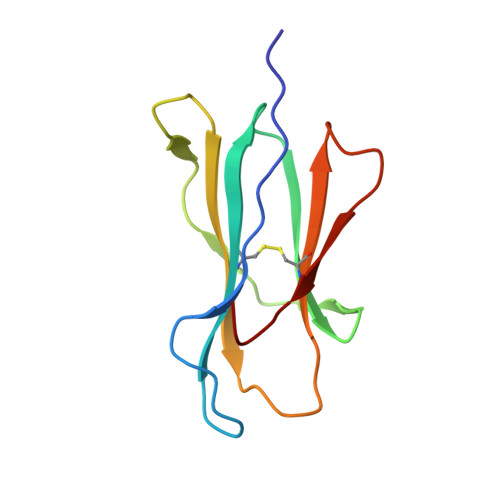
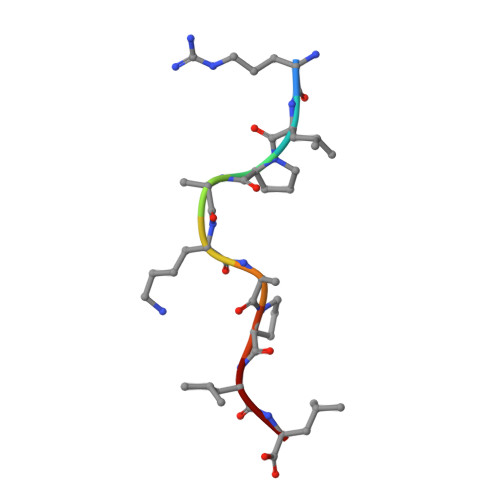
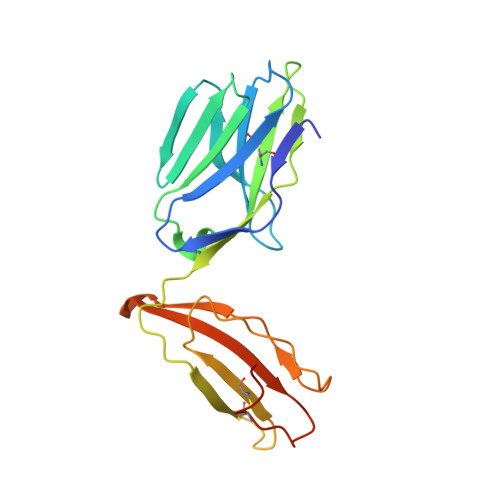
An HLA-E-targeted TCR bispecific molecule redirects T cell immunity against Mycobacterium tuberculosis.
Paterson, R.L., La Manna, M.P., Arena De Souza, V., Walker, A., Gibbs-Howe, D., Kulkarni, R., Fergusson, J.R., Mulakkal, N.C., Monteiro, M., Bunjobpol, W., Dembek, M., Martin-Urdiroz, M., Grant, T., Barber, C., Garay-Baquero, D.J., Tezera, L.B., Lowne, D., Britton-Rivet, C., Pengelly, R., Chepisiuk, N., Singh, P.K., Woon, A.P., Powlesland, A.S., McCully, M.L., Caccamo, N., Salio, M., Badami, G.D., Dorrell, L., Knox, A., Robinson, R., Elkington, P., Dieli, F., Lepore, M., Leonard, S., Godinho, L.F.(2024) Proc Natl Acad Sci U S A 121: e2318003121-e2318003121
- PubMed: 38691588
- DOI: https://doi.org/10.1073/pnas.2318003121
- Primary Citation of Related Structures:
8QFY - PubMed Abstract:
Peptides presented by HLA-E, a molecule with very limited polymorphism, represent attractive targets for T cell receptor (TCR)-based immunotherapies to circumvent the limitations imposed by the high polymorphism of classical HLA genes in the human population. Here, we describe a TCR-based bispecific molecule that potently and selectively binds HLA-E in complex with a peptide encoded by the inhA gene of Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis in humans. We reveal the biophysical and structural bases underpinning the potency and specificity of this molecule and demonstrate its ability to redirect polyclonal T cells to target HLA-E-expressing cells transduced with mycobacterial inhA as well as primary cells infected with virulent Mtb. Additionally, we demonstrate elimination of Mtb-infected cells and reduction of intracellular Mtb growth. Our study suggests an approach to enhance host T cell immunity against Mtb and provides proof of principle for an innovative TCR-based therapeutic strategy overcoming HLA polymorphism and therefore applicable to a broader patient population.
- Immunocore Ltd., Abingdon, Oxfordshire OX14 4RY, United Kingdom.
Organizational Affiliation: