PKR kinase domain- eIF2alpha in complex with compound
Nawrotek, A., Vuillard, L., Miallau, L.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Eukaryotic translation initiation factor 2 subunit alpha | 173 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: SUI2, TIF211, YJR007W, J1429 | 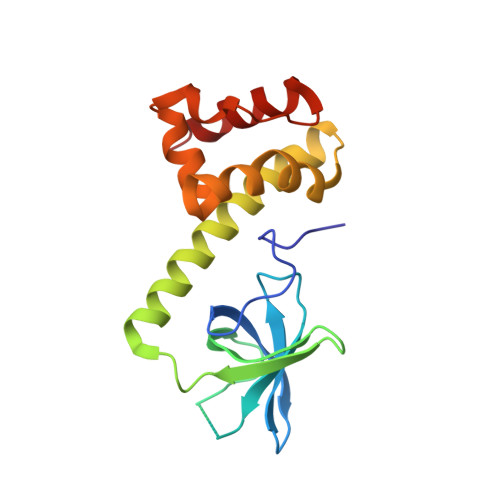 | |
UniProt | |||||
Find proteins for P20459 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P20459 Go to UniProtKB: P20459 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P20459 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Interferon-induced, double-stranded RNA-activated protein kinase | 273 | Homo sapiens | Mutation(s): 0 Gene Names: EIF2AK2, PKR, PRKR EC: 2.7.11.1 (PDB Primary Data), 2.7.10.2 (PDB Primary Data) | 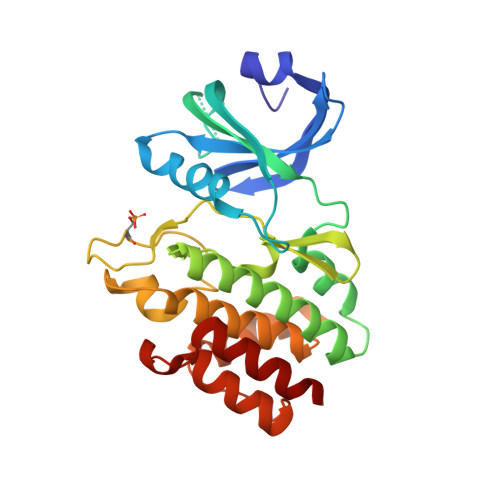 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P19525 (Homo sapiens) Explore P19525 Go to UniProtKB: P19525 | |||||
PHAROS: P19525 GTEx: ENSG00000055332 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P19525 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| UH3 (Subject of Investigation/LOI) Query on UH3 | C [auth B] | (3~{Z})-3-[(4-methyl-1~{H}-imidazol-5-yl)methylidene]-2-oxidanylidene-1~{H}-indole-5-carboxamide C14 H12 N4 O2 NQRCNELXESTXEP-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| TPO Query on TPO | B | L-PEPTIDE LINKING | C4 H10 N O6 P |  | THR |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 85.028 | α = 90 |
| b = 85.028 | β = 90 |
| c = 168.94 | γ = 120 |
| Software Name | Purpose |
|---|---|
| BUSTER | refinement |
| autoPROC | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |