Development of an active-site titrant for SARS-CoV-2 main protease as an indispensable tool for evaluating enzyme kinetics.
Voget, R., Breidenbach, J., Claff, T., Hingst, A., Sylvester, K., Steinebach, C., Vu, L.P., Weisse, R.H., Bartz, U., Strater, N., Muller, C.E., Gutschow, M.(2024) Acta Pharm Sin B 14: 2349-2357
- PubMed: 38799620
- DOI: https://doi.org/10.1016/j.apsb.2024.03.001
- Primary Citation of Related Structures:
8QDC - PubMed Abstract:
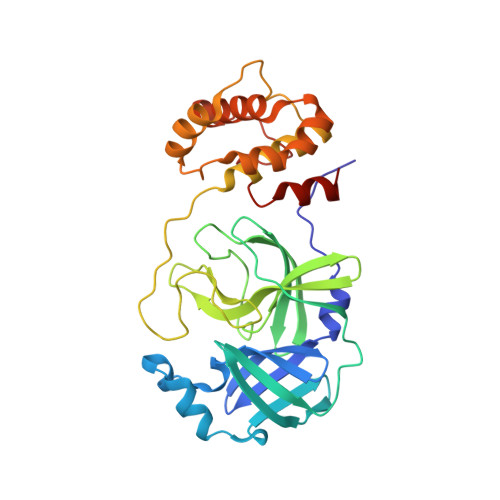
A titrant for the SARS-CoV-2 main protease (M pro ) was developed that enables, for the first time, the exact determination of the concentration of the enzymatically active M pro by active-site titration. The covalent binding mode of the tetrapeptidic titrant was elucidated by the determination of the crystal structure of the enzyme-titrant complex. Four fluorogenic substrates of M pro , including a prototypical, internally quenched Dabcyl-EDANS peptide, were compared in terms of solubility under typical assay conditions. By exploiting the new titrant, key kinetic parameters for the M pro -catalyzed cleavage of these substrates were determined.
Organizational Affiliation:
Pharmaceutical Institute, Pharmaceutical & Medicinal Chemistry, University of Bonn, Bonn 53121, Germany.