Expanding the Anti-Phl p 7 Antibody Toolkit: An Anti-Idiotype Nanobody Inhibitor.
Vester, S.K., Davies, A.M., Beavil, R.L., Sandhar, B.S., Beavil, A.J., Gould, H.J., Sutton, B.J., McDonnell, J.M.(2023) Antibodies (Basel) 12
- PubMed: 37987253
- DOI: https://doi.org/10.3390/antib12040075
- Primary Citation of Related Structures:
8Q6K - PubMed Abstract:
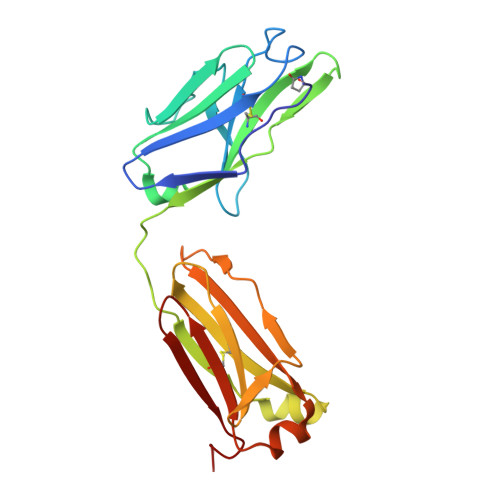
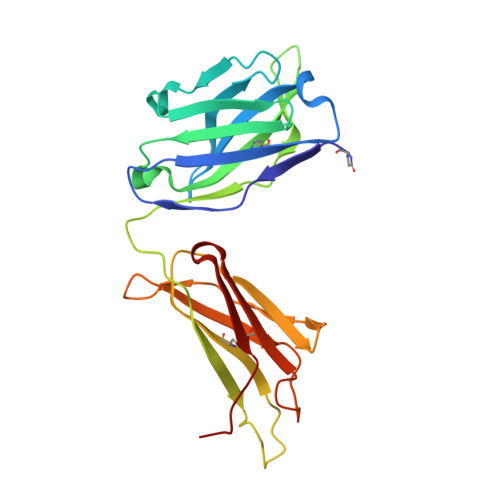
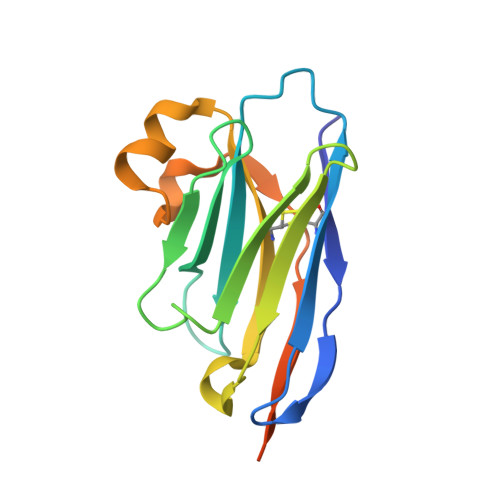
We have previously produced a toolkit of antibodies, comprising recombinant human antibodies of all but one of the human isotypes, directed against the polcalcin family antigen Phl p 7. In this work, we complete the toolkit of human antibody isotypes with the IgD version of the anti-Phl p 7 monoclonal antibody. We also raised a set of nanobodies against the IgD anti-Phl p 7 antibody and identify and characterize one paratope-specific nanobody. This nanobody also binds to the IgE isotype of this antibody, which shares the same idiotype, and orthosterically inhibits the interaction with Phl p 7. The 2.1 Å resolution X-ray crystal structure of the nanobody in complex with the IgD Fab is described.
- Randall Centre for Cell and Molecular Biophysics, King's College London, New Hunt's House, London SE1 1UL, UK.
Organizational Affiliation: