HIF prolyl hydroxylase 2 in complex with HIF2alpha-CODD
Fiorini, G., Figg Jr, W.D., Schofield, C.J.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Egl nine homolog 1 | 233 | Homo sapiens | Mutation(s): 0 Gene Names: EGLN1, C1orf12, PNAS-118, PNAS-137 EC: 1.14.11.29 | 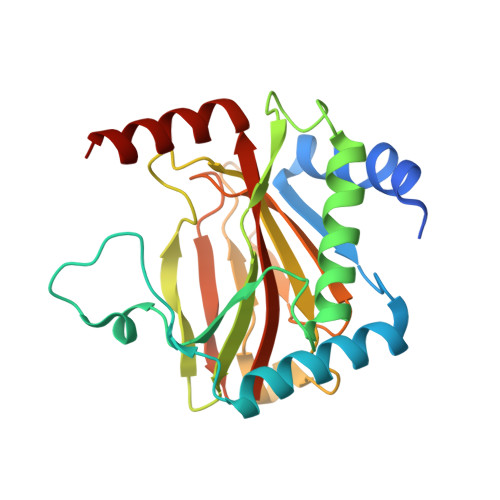 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9GZT9 (Homo sapiens) Explore Q9GZT9 Go to UniProtKB: Q9GZT9 | |||||
PHAROS: Q9GZT9 GTEx: ENSG00000135766 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9GZT9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Endothelial PAS domain-containing protein 1 | 20 | Homo sapiens | Mutation(s): 0 Gene Names: EPAS1, BHLHE73, HIF2A, MOP2, PASD2 | 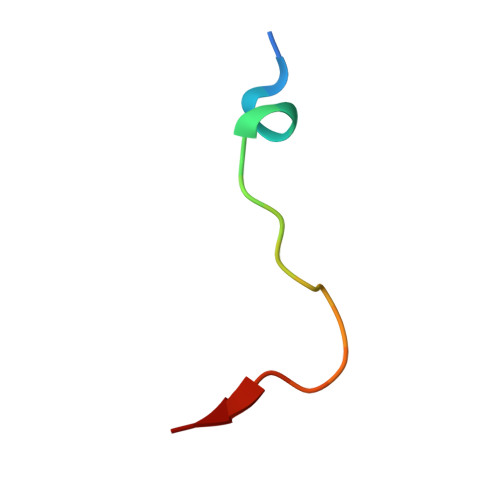 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q99814 (Homo sapiens) Explore Q99814 Go to UniProtKB: Q99814 | |||||
PHAROS: Q99814 GTEx: ENSG00000116016 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q99814 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AKG (Subject of Investigation/LOI) Query on AKG | I [auth A] | 2-OXOGLUTARIC ACID C5 H6 O5 KPGXRSRHYNQIFN-UHFFFAOYSA-N |  | ||
| FE (Subject of Investigation/LOI) Query on FE | H [auth A] | FE (III) ION Fe VTLYFUHAOXGGBS-UHFFFAOYSA-N |  | ||
| CL Query on CL | F [auth A], G [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | C [auth A], D [auth A], E [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 130.9 | α = 90 |
| b = 38.12 | β = 90 |
| c = 42.77 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| DIALS | data scaling |
| xia2 | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | -- |
| University of Oxford, Medical Sciences Internal Fund (MSIF) | United Kingdom | -- |
| Cancer Research UK | United Kingdom | -- |
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | -- |