Structural analysis of the BisI family of modification dependent restriction endonucleases.
Szafran, K., Rafalski, D., Skowronek, K., Wojciechowski, M., Kazrani, A.A., Gilski, M., Xu, S.Y., Bochtler, M.(2024) Nucleic Acids Res 52: 9103-9118
- PubMed: 39041409
- DOI: https://doi.org/10.1093/nar/gkae634
- Primary Citation of Related Structures:
8Q5M, 8Q5N, 8Q5O, 8RPX - PubMed Abstract:
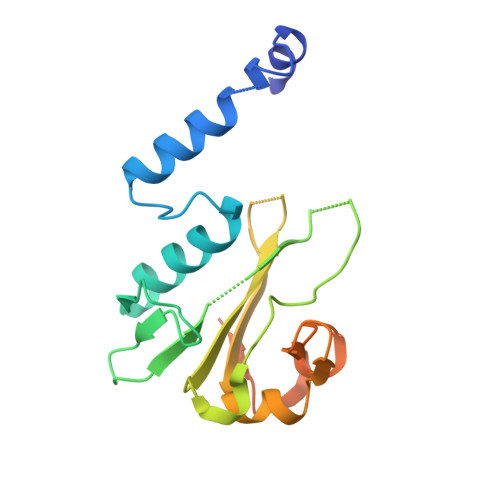
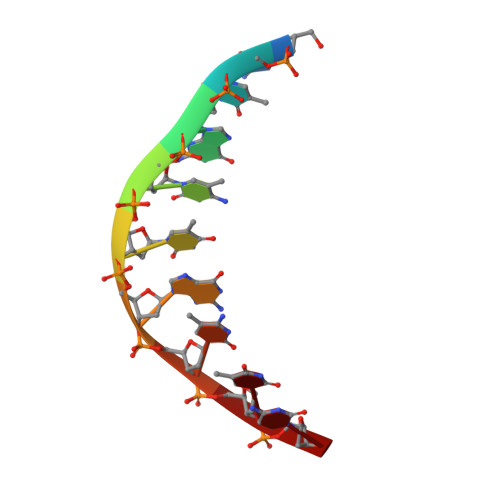
The BisI family of restriction endonucleases is unique in requiring multiple methylated or hydroxymethylated cytosine residues within a short recognition sequence (GCNGC), and in cleaving directly within this sequence, rather than at a distance. Here, we report that the number of modified cytosines that are required for cleavage can be tuned by the salt concentration. We present crystal structures of two members of the BisI family, NhoI and Eco15I_Ntd (N-terminal domain of Eco15I), in the absence of DNA and in specific complexes with tetra-methylated GCNGC target DNA. The structures show that NhoI and Eco15I_Ntd sense modified cytosine bases in the context of double-stranded DNA (dsDNA) without base flipping. In the co-crystal structures of NhoI and Eco15I_Ntd with DNA, the internal methyl groups (G5mCNGC) interact with the side chains of an (H/R)(V/I/T/M) di-amino acid motif near the C-terminus of the distal enzyme subunit and arginine residue from the proximal subunit. The external methyl groups (GCNG5mC) interact with the proximal enzyme subunit, mostly through main chain contacts. Surface plasmon resonance analysis for Eco15I_Ntd shows that the internal and external methyl binding pockets contribute about equally to sensing of cytosine methyl groups.
Organizational Affiliation:
International Institute of Molecular and Cell Biology, Warsaw, Poland.