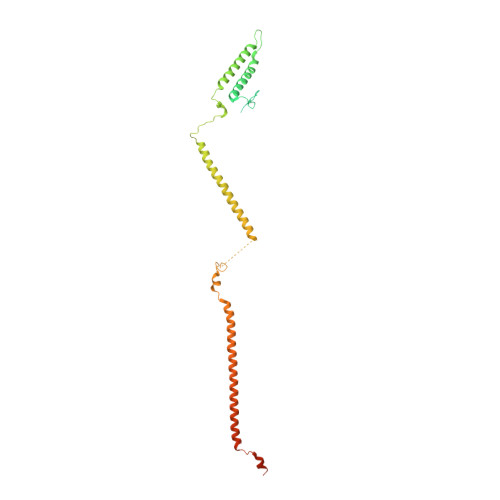
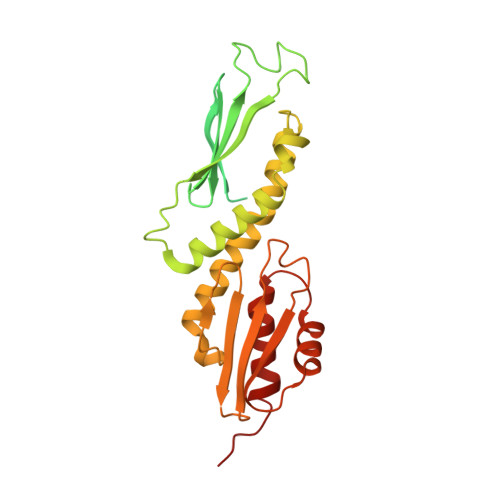
Structure of the human KMN complex and implications for regulation of its assembly.
Polley, S., Raisch, T., Ghetti, S., Korner, M., Terbeck, M., Grater, F., Raunser, S., Aponte-Santamaria, C., Vetter, I.R., Musacchio, A.(2024) Nat Struct Mol Biol 31: 861-873
- PubMed: 38459128
- DOI: https://doi.org/10.1038/s41594-024-01230-9
- Primary Citation of Related Structures:
8Q5H - PubMed Abstract:
Biorientation of chromosomes during cell division is necessary for precise dispatching of a mother cell's chromosomes into its two daughters. Kinetochores, large layered structures built on specialized chromosome loci named centromeres, promote biorientation by binding and sensing spindle microtubules. One of the outer layer main components is a ten-subunit assembly comprising Knl1C, Mis12C and Ndc80C (KMN) subcomplexes. The KMN is highly elongated and docks on kinetochores and microtubules through interfaces at its opposite extremes. Here, we combine cryogenic electron microscopy reconstructions and AlphaFold2 predictions to generate a model of the human KMN that reveals all intra-KMN interfaces. We identify and functionally validate two interaction interfaces that link Mis12C to Ndc80C and Knl1C. Through targeted interference experiments, we demonstrate that this mutual organization strongly stabilizes the KMN assembly. Our work thus reports a comprehensive structural and functional analysis of this part of the kinetochore microtubule-binding machinery and elucidates the path of connections from the chromatin-bound components to the force-generating components.
- Department of Mechanistic Cell Biology, Max Planck Institute of Molecular Physiology, Dortmund, Germany.
Organizational Affiliation: