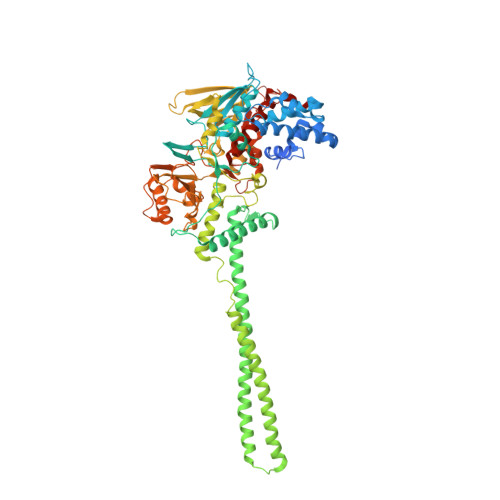
Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Lee, K., Barone, M., Waterbury, A.L., Jiang, H., Nam, E., DuBois-Coyne, S.E., Whedon, S.D., Wang, Z.A., Caroli, J., Neal, K., Ibeabuchi, B., Dhoondia, Z., Kuroda, M.I., Liau, B.B., Beck, S., Mattevi, A., Cole, P.A.(2025) Nat Chem Biol 21: 227-237
- PubMed: 38965385
- DOI: https://doi.org/10.1038/s41589-024-01671-9
- Primary Citation of Related Structures:
8Q1G, 8Q1H, 8Q1J - PubMed Abstract:
Biochemical crosstalk between two or more histone modifications is often observed in epigenetic enzyme regulation, but its functional significance in cells has been difficult to discern. Previous enzymatic studies revealed that Lys14 acetylation of histone H3 can inhibit Lys4 demethylation by lysine-specific demethylase 1 (LSD1). In the present study, we engineered a mutant form of LSD1, Y391K, which renders the nucleosome demethylase activity of LSD1 insensitive to Lys14 acetylation. K562 cells with the Y391K LSD1 CRISPR knockin show decreased expression of a set of genes associated with cellular adhesion and myeloid leukocyte activation. Chromatin profiling revealed that the cis-regulatory regions of these silenced genes display a higher level of H3 Lys14 acetylation, and edited K562 cells show diminished H3 mono-methyl Lys4 near these silenced genes, consistent with a role for enhanced LSD1 demethylase activity. These findings illuminate the functional consequences of disconnecting histone modification crosstalk for a key epigenetic enzyme.
- Division of Genetics, Department of Medicine, Brigham and Women's Hospital, Boston, MA, USA.
Organizational Affiliation: