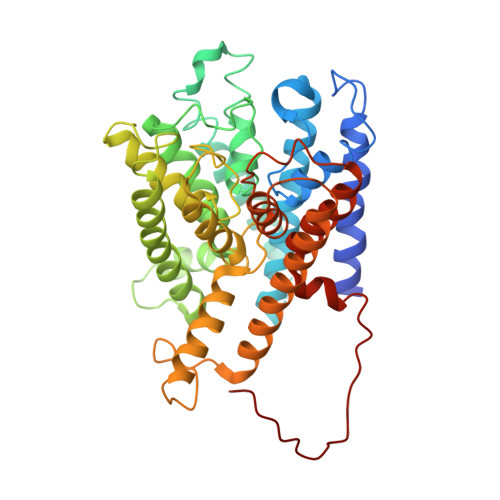
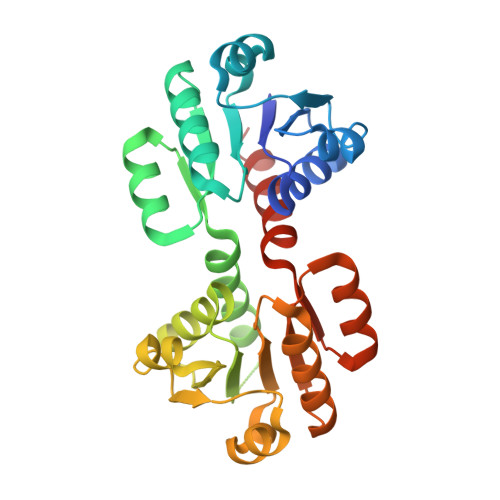
c-di-AMP determines the hierarchical organization of bacterial RCK proteins.
Rocha, R., Jorge, J.M.P., Teixeira-Duarte, C.M., Figueiredo-Costa, I.R., Cereija, T.B., Ferreira-Teixeira, P.F., Herzberg, C., Stulke, J., Morais-Cabral, J.H.(2024) Proc Natl Acad Sci U S A 121: e2318666121-e2318666121
- PubMed: 38652747
- DOI: https://doi.org/10.1073/pnas.2318666121
- Primary Citation of Related Structures:
8POO - PubMed Abstract:
In bacteria, intracellular K + is involved in the regulation of membrane potential, cytosolic pH, and cell turgor as well as in spore germination, environmental adaptation, cell-to-cell communication in biofilms, antibiotic sensitivity, and infectivity. The second messenger cyclic-di-AMP (c-di-AMP) has a central role in modulating the intracellular K + concentration in many bacterial species, controlling transcription and function of K + channels and transporters. However, our understanding of how this regulatory network responds to c-di-AMP remains poor. We used the RCK (Regulator of Conductance of K + ) proteins that control the activity of Ktr channels in Bacillus subtilis as a model system to analyze the regulatory function of c-di-AMP with a combination of in vivo and in vitro functional and structural characterization. We determined that the two RCK proteins (KtrA and KtrC) are neither physiologically redundant or functionally equivalent. KtrC is the physiologically dominant RCK protein in the regulation of Ktr channel activity. In explaining this hierarchical organization, we found that, unlike KtrA, KtrC is very sensitive to c-di-AMP inactivation and lack of c-di-AMP regulation results in RCK protein toxicity, most likely due to unregulated K + flux. We also found that KtrC can assemble with KtrA, conferring c-di-AMP regulation to the functional KtrA/KtrC heteromers and potentially compensating KtrA toxicity. Altogether, we propose that the central role of c-di-AMP in the control of the K + machinery, by modulating protein levels through gene transcription and by regulating protein activity, has determined the evolutionary selection of KtrC as the dominant RCK protein, shaping the hierarchical organization of regulatory components of the K + machinery.
- Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Porto 4200-135, Portugal.
Organizational Affiliation: