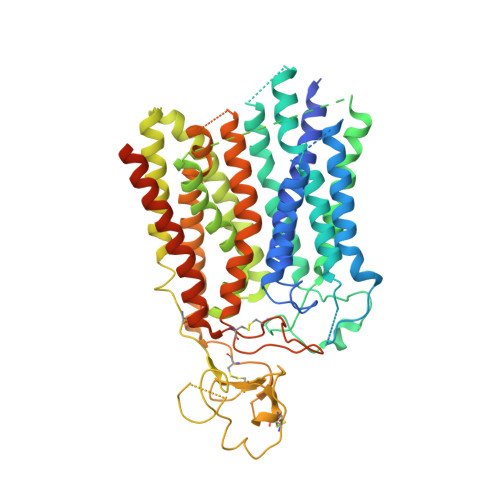
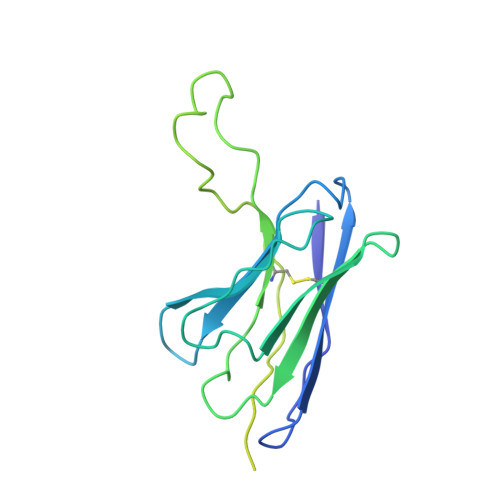
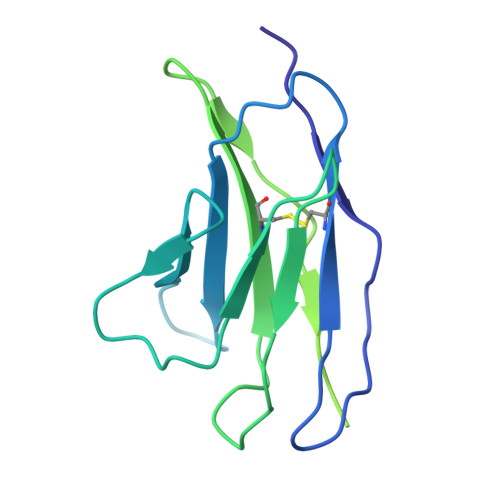
Structure of human drug transporters OATP1B1 and OATP1B3.
Ciuta, A.D., Nosol, K., Kowal, J., Mukherjee, S., Ramirez, A.S., Stieger, B., Kossiakoff, A.A., Locher, K.P.(2023) Nat Commun 14: 5774-5774
- PubMed: 37723174
- DOI: https://doi.org/10.1038/s41467-023-41552-8
- Primary Citation of Related Structures:
8PG0, 8PHW - PubMed Abstract:
The organic anion transporting polypeptides OATP1B1 and OATP1B3 are membrane proteins that mediate uptake of drugs into the liver for subsequent conjugation and biliary excretion, a key step in drug elimination from the human body. Polymorphic variants of these transporters can cause reduced drug clearance and adverse drug effects such as statin-induced rhabdomyolysis, and co-administration of OATP substrates can lead to damaging drug-drug interaction. Despite their clinical relevance in drug disposition and pharmacokinetics, the structure and mechanism of OATPs are unknown. Here we present cryo-EM structures of human OATP1B1 and OATP1B3 bound to synthetic Fab fragments and in functionally distinct states. A single estrone-3-sulfate molecule is bound in a pocket located in the C-terminal half of OATP1B1. The shape and chemical nature of the pocket rationalize the preference for diverse organic anions and allow in silico docking of statins. The structure of OATP1B3 is determined in a drug-free state but reveals a bicarbonate molecule bound to the conserved signature motif and a histidine residue that is prevalent in OATPs exhibiting pH-dependent activity.
- Institute of Molecular Biology and Biophysics, Department of Biology, ETH Zürich, Zürich, Switzerland.
Organizational Affiliation: