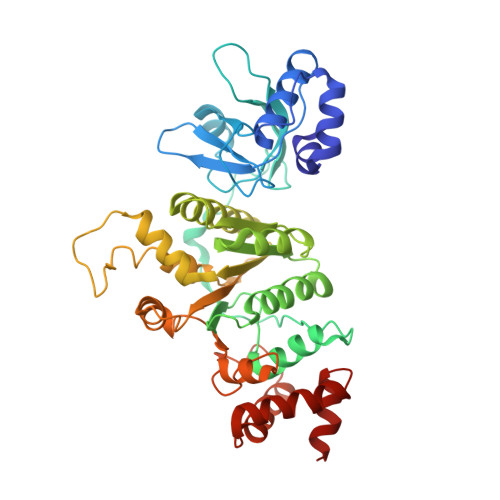
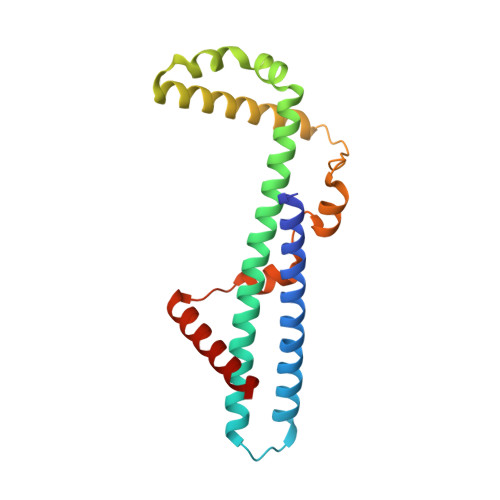
The Psu protein of phage satellite P4 inhibits transcription termination factor rho by forced hyper-oligomerization.
Gjorgjevikj, D., Kumar, N., Wang, B., Hilal, T., Said, N., Loll, B., Artsimovitch, I., Sen, R., Wahl, M.C.(2025) Nat Commun 16: 550-550
- PubMed: 39788982
- DOI: https://doi.org/10.1038/s41467-025-55897-9
- Primary Citation of Related Structures:
8PEU, 8PEW, 8PEX, 8PEY, 9GCS, 9GCT, 9GCU - PubMed Abstract:
Many bacteriophages modulate host transcription to favor expression of their own genomes. Phage satellite P4 polarity suppression protein, Psu, a building block of the viral capsid, inhibits hexameric transcription termination factor, ρ, by presently unknown mechanisms. Our cryogenic electron microscopy structures of ρ-Psu complexes show that Psu dimers clamp two inactive, open ρ rings and promote their expansion to higher-oligomeric states. ATPase, nucleotide binding and nucleic acid binding studies revealed that Psu hinders ρ ring closure and traps nucleotides in their binding pockets on ρ. Structure-guided mutagenesis in combination with growth, pull-down, and termination assays further delineated the functional ρ-Psu interfaces in vivo. Bioinformatic analyses revealed that Psu is associated with a wide variety of phage defense systems across Enterobacteriaceae, suggesting that Psu may regulate expression of anti-phage genes. Our findings show that modulation of the ρ oligomeric state via diverse strategies is a pervasive gene regulatory principle in bacteria.
- Laboratory of Structural Biochemistry, Institute of Chemistry and Biochemistry, Freie Universität Berlin, Berlin, Germany.
Organizational Affiliation: