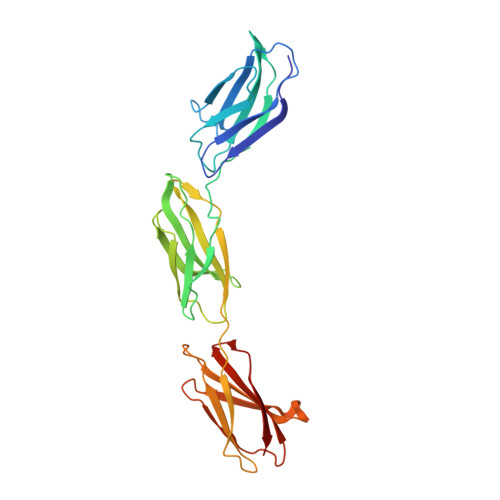
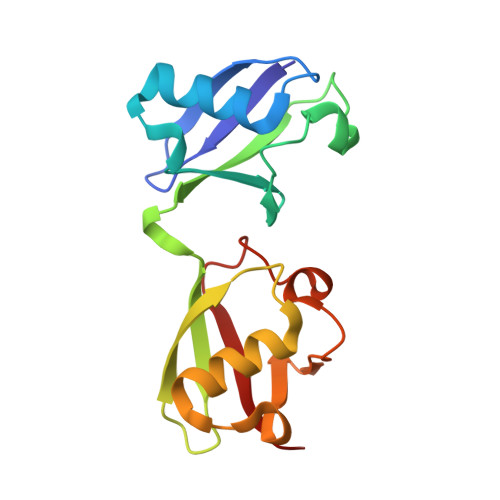
Ubiquitin-derived artificial binding proteins targeting oncofetal fibronectin reveal scaffold plasticity by beta-strand slippage.
Katzschmann, A., Haupts, U., Reimann, A., Settele, F., Gloser-Braunig, M., Fiedler, E., Parthier, C.(2024) Commun Biol 7: 907-907
- PubMed: 39068227
- DOI: https://doi.org/10.1038/s42003-024-06569-9
- Primary Citation of Related Structures:
8PEQ, 8PF0 - PubMed Abstract:
Affilin proteins, artificial binding proteins based on the ubiquitin scaffold, have been generated by directed protein evolution to yield de-novo variants that bind the extra-domain B (EDB) of oncofetal fibronectin, an established marker of tumor neovasculature. The crystal structures of two EDB-specific Affilin variants reveal a striking structural plasticity of the ubiquitin scaffold, characterised by β-strand slippage, leading to different negative register shifts of the β5 strands. This process recruits amino acid residues from β5 towards the N-terminus to an adjacent loop region and subsequent residues into β5, respectively, remodeling the binding interface and leading to target specificity and affinity. Protein backbone alterations resulting from β-strand register shifts, as seen in the ubiquitin fold, can pose additional challenges to protein engineering as structural evidence of these events is still limited and they are difficult to predict. However, they can surface under the selection pressure of directed evolution and suggest that backbone plasticity allowing β-strand slippages can increase structural diversity, enhancing the evolutionary potential of a protein scaffold.
- Navigo Proteins GmbH, Heinrich-Damerow-Straße 1, 06120, Halle (Saale), Germany.
Organizational Affiliation: