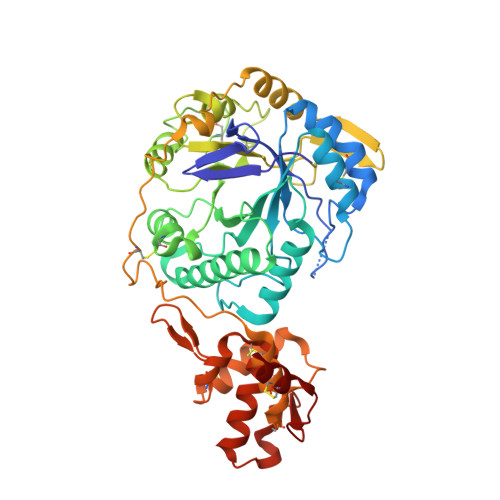
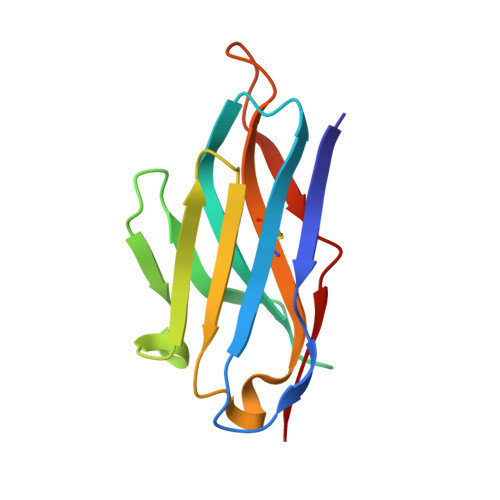
Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Redrado-Hernandez, S., Macias-Leon, J., Castro-Lopez, J., Belen Sanz, A., Dolader, E., Arias, M., Gonzalez-Ramirez, A.M., Sanchez-Navarro, D., Petryk, Y., Farkas, V., Vincke, C., Muyldermans, S., Garcia-Barbazan, I., Del Agua, C., Zaragoza, O., Arroyo, J., Pardo, J., Galvez, E.M., Hurtado-Guerrero, R.(2024) Angew Chem Int Ed Engl 63: e202405823-e202405823
- PubMed: 38856634
- DOI: https://doi.org/10.1002/anie.202405823
- Primary Citation of Related Structures:
8PE1, 8PE2 - PubMed Abstract:
Invasive fungal disease accounts for ~3.8 million deaths annually, an unacceptable rate that urgently prompts the discovery of new knowledge-driven treatments. We report the use of camelid single-domain nanobodies (Nbs) against fungal β-1,3-glucanosyltransferases (Gel) involved in β-1,3-glucan transglycosylation. Crystal structures of two Nbs with Gel4 from Aspergillus fumigatus revealed binding to a dissimilar CBM43 domain and a highly conserved catalytic domain across fungal species, respectively. Anti-Gel4 active site Nb3 showed significant antifungal efficacy in vitro and in vivo prophylactically and therapeutically against different A. fumigatus and Cryptococcus neoformans isolates, reducing the fungal burden and disease severity, thus significantly improving immunocompromised animal survival. Notably, C. deneoformans (serotype D) strains were more susceptible to Nb3 and genetic Gel deletion than C. neoformans (serotype A) strains, indicating a key role for β-1,3-glucan remodelling in C. deneoformans survival. These findings add new insights about the role of b-1,3-glucan in fungal biology and demonstrate the potential of nanobodies in targeting fungal enzymes to combat invasive fungal diseases.
- Instituto de Carboquimica, Instituto de Carboquimica, SPAIN.
Organizational Affiliation: