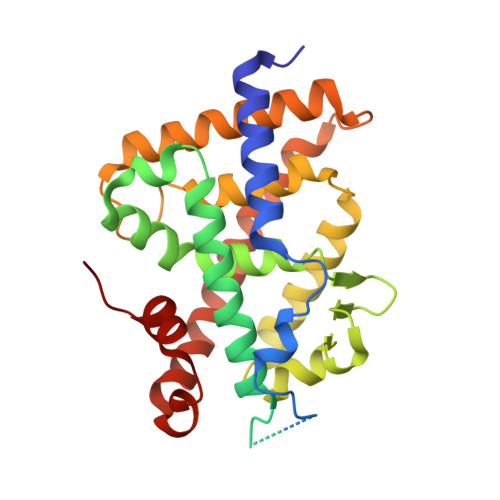
A vitamin D-based strategy overcomes chemoresistance in prostate cancer.
Len-Tayon, K., Beraud, C., Fauveau, C., Belorusova, A.Y., Chebaro, Y., Mourino, A., Massfelder, T., Chauchereau, A., Metzger, D., Rochel, N., Laverny, G.(2024) Br J Pharmacol 181: 4279-4293
- PubMed: 38982588
- DOI: https://doi.org/10.1111/bph.16492
- Primary Citation of Related Structures:
8P9W, 8P9X - PubMed Abstract:
Castration-resistant prostate cancer (CRPC) is a common male malignancy that requires new therapeutic strategies due to acquired resistance to its first-line treatment, docetaxel. The benefits of vitamin D on prostate cancer (PCa) progression have been previously reported. This study aimed to investigate the effects of vitamin D on chemoresistance in CRPC. Structure function relationships of potent vitamin D analogues were determined. The combination of the most potent analogue and docetaxel was explored in chemoresistant primary PCa spheroids and in a xenograft mouse model derived from a patient with a chemoresistant CRPC. Here, we show that Xe4MeCF3 is more potent than the natural ligand to induce vitamin D receptor (VDR) transcriptional activities and that it has a larger therapeutic window. Moreover, we demonstrate that VDR agonists restore docetaxel sensitivity in PCa spheroids. Importantly, Xe4MeCF3 reduces tumour growth in a chemoresistant CRPC patient-derived xenograft. In addition, this treatment targets signalling pathways associated with cancer progression in the remaining cells. Taken together, these results unravel the potency of VDR agonists to overcome chemoresistance in CRPC and open new avenues for the clinical management of PCa.
- Institute of Genetics and Molecular and Cellular Biology (IGBMC), Illkirch-Graffenstaden, France.
Organizational Affiliation: