Pyridylpiperazine efflux pump inhibitor boosts in vivo antibiotic efficacy against K. pneumoniae.
Vieira Da Cruz, A., Jimenez-Castellanos, J.C., Bornsen, C., Van Maele, L., Compagne, N., Pradel, E., Muller, R.T., Meurillon, V., Soulard, D., Piveteau, C., Biela, A., Dumont, J., Leroux, F., Deprez, B., Willand, N., Pos, K.M., Frangakis, A.S., Hartkoorn, R.C., Flipo, M.(2024) EMBO Mol Med 16: 93-111
- PubMed: 38177534
- DOI: https://doi.org/10.1038/s44321-023-00007-9
- Primary Citation of Related Structures:
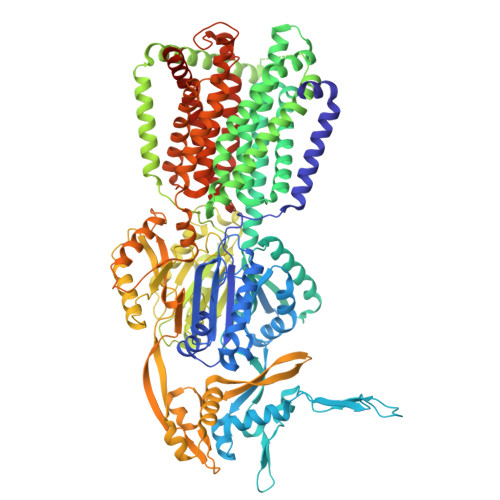
8P1I - PubMed Abstract:
Antimicrobial resistance is a global problem, rendering conventional treatments less effective and requiring innovative strategies to combat this growing threat. The tripartite AcrAB-TolC efflux pump is the dominant constitutive system by which Enterobacterales like Escherichia coli and Klebsiella pneumoniae extrude antibiotics. Here, we describe the medicinal chemistry development and drug-like properties of BDM91288, a pyridylpiperazine-based AcrB efflux pump inhibitor. In vitro evaluation of BDM91288 confirmed it to potentiate the activity of a panel of antibiotics against K. pneumoniae as well as revert clinically relevant antibiotic resistance mediated by acrAB-tolC overexpression. Using cryo-EM, BDM91288 binding to the transmembrane region of K. pneumoniae AcrB was confirmed, further validating the mechanism of action of this inhibitor. Finally, proof of concept studies demonstrated that oral administration of BDM91288 significantly potentiated the in vivo efficacy of levofloxacin treatment in a murine model of K. pneumoniae lung infection.
- Univ. Lille, Inserm, Institut Pasteur de Lille, U1177 - Drugs and Molecules for Living Systems, F-59000, Lille, France.
Organizational Affiliation: