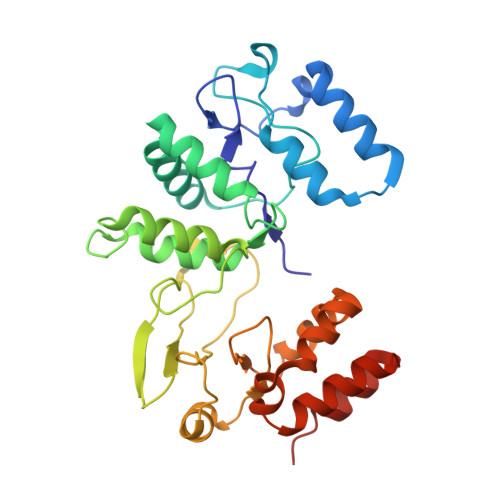
Cryo-EM structure of rotavirus B NSP2 reveals its unique tertiary architecture.
Chamera, S., Wycisk, K., Czarnocki-Cieciura, M., Nowotny, M.(2024) J Virol 98: e0166023-e0166023
- PubMed: 38421167
- DOI: https://doi.org/10.1128/jvi.01660-23
- Primary Citation of Related Structures:
8P00 - PubMed Abstract:
Rotavirus (RV) NSP2 is a multifunctional RNA chaperone that exhibits numerous activities that are essential for replication and viral genome packaging. We performed an in silico analysis that highlighted a distant relationship of NSP2 from rotavirus B (RVB) to proteins from other human RVs. We solved a cryo-electron microscopy structure of RVB NSP2 that shows structural differences with corresponding proteins from other human RVs. Based on the structure, we identified amino acid residues that are involved in RNA interactions. Anisotropy titration experiments showed that these residues are important for nucleic acid binding. We also identified structural motifs that are conserved in all RV species. Collectively, our data complete the structural characterization of rotaviral NSP2 protein and demonstrate its structural diversity among RV species.IMPORTANCERotavirus B (RVB), also known as adult diarrhea rotavirus, has caused epidemics of severe diarrhea in China, India, and Bangladesh. Thousands of people are infected in a single RVB epidemic. However, information on this group of rotaviruses remains limited. As NSP2 is an essential protein in the viral life cycle, including its role in the formation of replication factories, it may be a target for future antiviral strategy against viruses with similar mechanisms.
- Laboratory of Protein Structure, International Institute of Molecular and Cell Biology, Warsaw, Poland.
Organizational Affiliation: