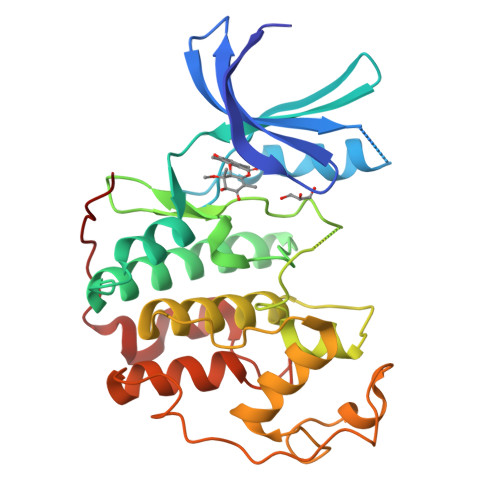
Resistance gene-guided genome mining reveals the roseopurpurins as inhibitors of cyclin-dependent kinases.
Dunbar, K.L., Perlatti, B., Liu, N., Cornelius, A., Mummau, D., Chiang, Y.M., Hon, L., Nimavat, M., Pallas, J., Kordes, S., Ng, H.L., Harvey, C.J.B.(2023) Proc Natl Acad Sci U S A 120: e2310522120-e2310522120
- PubMed: 37983497
- DOI: https://doi.org/10.1073/pnas.2310522120
- Primary Citation of Related Structures:
8OY2 - PubMed Abstract:
With the significant increase in the availability of microbial genome sequences in recent years, resistance gene-guided genome mining has emerged as a powerful approach for identifying natural products with specific bioactivities. Here, we present the use of this approach to reveal the roseopurpurins as potent inhibitors of cyclin-dependent kinases (CDKs), a class of cell cycle regulators implicated in multiple cancers. We identified a biosynthetic gene cluster (BGC) with a putative resistance gene with homology to human CDK2. Using targeted gene disruption and transcription factor overexpression in Aspergillus uvarum , and heterologous expression of the BGC in Aspergillus nidulans, we demonstrated that roseopurpurin C ( 1 ) is produced by this cluster and characterized its biosynthesis. We determined the potency, specificity, and mechanism of action of 1 as well as multiple intermediates and shunt products produced from the BGC. We show that 1 inhibits human CDK2 with a K iapp of 44 nM, demonstrates selectivity for clinically relevant members of the CDK family, and induces G1 cell cycle arrest in HCT116 cells. Structural analysis of 1 complexed with CDK2 revealed the molecular basis of ATP-competitive inhibition.
Organizational Affiliation:
Hexagon Bio, Menlo Park, CA 94025.