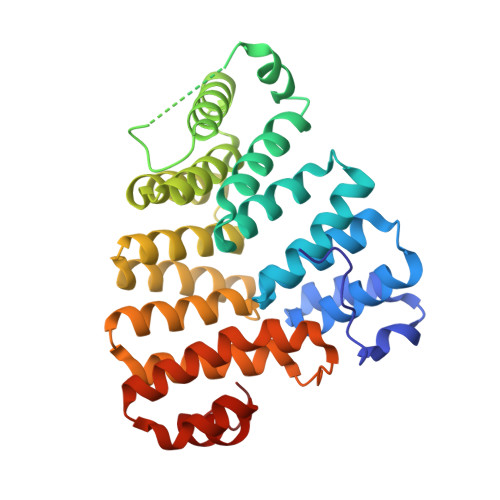
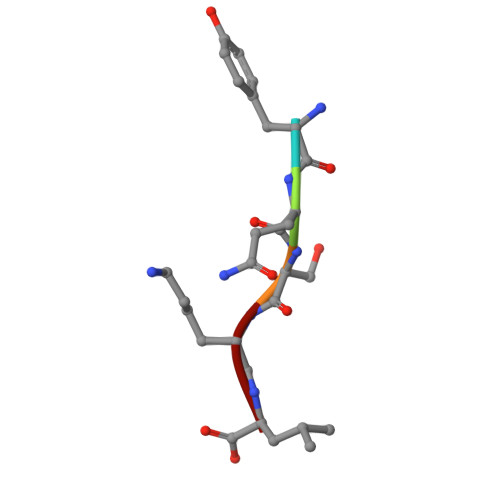
Structural dynamics of the TPR domain of the peroxisomal cargo receptor Pex5 in Trypanosoma.
Banasik, M., Napolitano, V., Blat, A., Abdulkarim, K., Plewka, J., Czaplewski, C., Gieldon, A., Kozak, M., Wladyka, B., Popowicz, G., Dubin, G.(2024) Int J Biol Macromol 280: 135510-135510
- PubMed: 39304044
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.135510
- Primary Citation of Related Structures:
8OS1, 9F8W - PubMed Abstract:
Peroxisomal protein import has been identified as a valid target in trypanosomiases, an important health threat in Central and South America. The importomer is built of multiple peroxins (Pex) and structural characterization of these proteins facilitates rational inhibitor development. We report crystal structures of the Trypanosoma brucei and T. cruzi tetratricopeptide repeat domain (TPR) of the cytoplasmic peroxisomal targeting signal 1 (PTS1) receptor Pex5. The structure of the TPR domain of TbPex5 represents an apo-form of the receptor which, together with the previously determined structure of the complex of TbPex5 TPR and PTS1 demonstrate significant receptor dynamics associated with signal peptide recognition. The structure of the complex of TPR domain of TcPex5 with PTS1 provided in this study details the molecular interactions that guide signal peptide recognition at the atomic level in the pathogenic species currently perceived as the most relevant among Trypanosoma. Small - angle X - ray scattering (SAXS) data obtained in solution supports the crystallographic findings on the compaction of the TPR domains of TbPex5 and TcPex5 upon interaction with the cargo.
- Malopolska Centre of Biotechnology, Jagiellonian University, Gronostajowa 7a, 30-387 Krakow, Poland.
Organizational Affiliation: