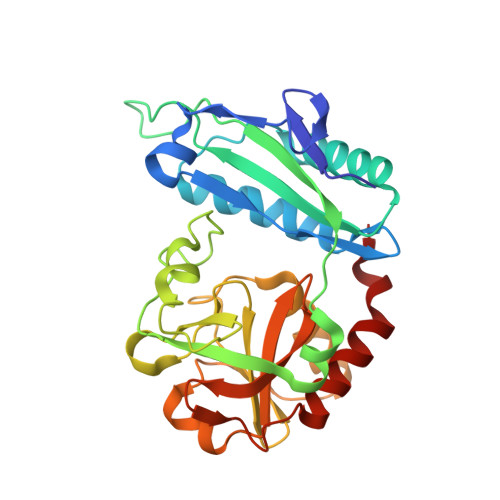
Probing of the structural and catalytic roles of the residues in the active site of transaminase from Aminobacterium colombiense
Shilova, S.A., Khrenova, M.G., Matyuta, I.O., Nikolaeva, A.Y., Klyachko, N.L., Minyaev, M.E., Boyko, K.M., Popov, V.O., Bezsudnova, E.Y.To be published.