Implications of Protein Interaction in the Speciation of Potential V IV O-Pyridinone Drugs.
Ferraro, G., Paolillo, M., Sciortino, G., Pisanu, F., Garribba, E., Merlino, A.(2023) Inorg Chem 62: 8407-8417
- PubMed: 37195003
- DOI: https://doi.org/10.1021/acs.inorgchem.3c01041
- Primary Citation of Related Structures:
8OM8, 8OMS, 8OMT - PubMed Abstract:
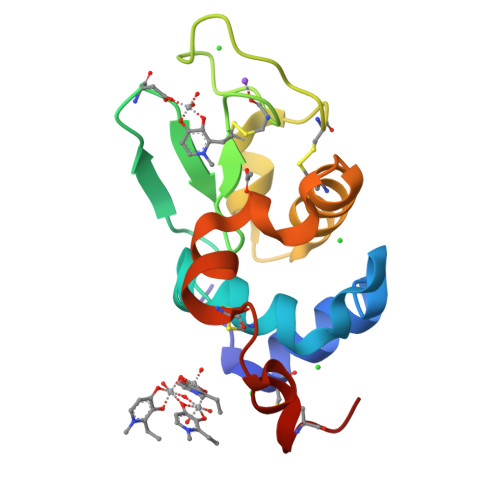
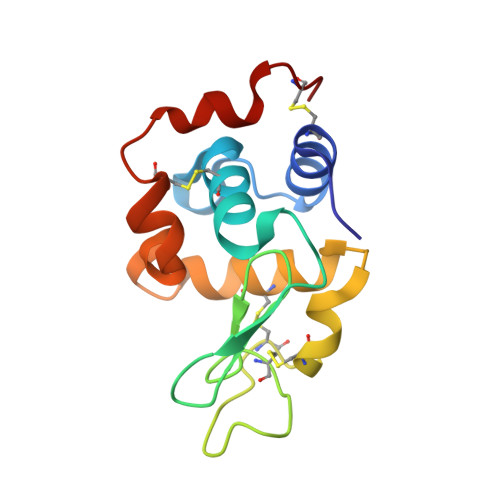
Vanadium complexes (VCs) are promising agents for the treatment, among others, of diabetes and cancer. The development of vanadium-based drugs is mainly limited by a scarce knowledge of the active species in the target organs, which is often determined by the interaction of VCs with biological macromolecules like proteins. Here, we have studied the binding of [V IV O(empp) 2 ] (where Hempp is 1-methyl-2-ethyl-3-hydroxy-4(1 H )-pyridinone), an antidiabetic and anticancer VC, with the model protein hen egg white lysozyme (HEWL) by electrospray ionization-mass spectrometry (ESI-MS), electron paramagnetic resonance (EPR), and X-ray crystallography. ESI-MS and EPR techniques reveal that, in aqueous solution, both the species [V IV O(empp) 2 ] and [V IV O(empp)(H 2 O)] + , derived from the first one upon the loss of a empp(-) ligand, interact with HEWL. Crystallographic data, collected under different experimental conditions, show covalent binding of [V IV O(empp)(H 2 O)] + to the side chain of Asp48, and noncovalent binding of cis -[V IV O(empp) 2 (H 2 O)], [V IV O(empp)(H 2 O)] + , [V IV O(empp)(H 2 O) 2 ] + , and of an unusual trinuclear oxidovanadium(V) complex, [V V 3 O 6 (empp) 3 (H 2 O)], with accessible sites on the protein surface. The possibility of covalent and noncovalent binding with different strength and of interaction with various sites favor the formation of adducts with the multiple binding of vanadium moieties, allowing the transport in blood and cellular fluids of more than one metal-containing species with a possible amplification of the biological effects.
Organizational Affiliation:
Department of Chemical Sciences, University of Naples Federico II, Complesso Universitario di Monte Sant'Angelo, Via Cintia, I-80126 Napoli, Italy.