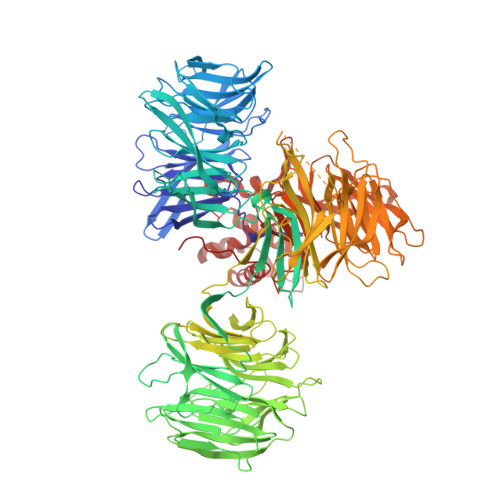
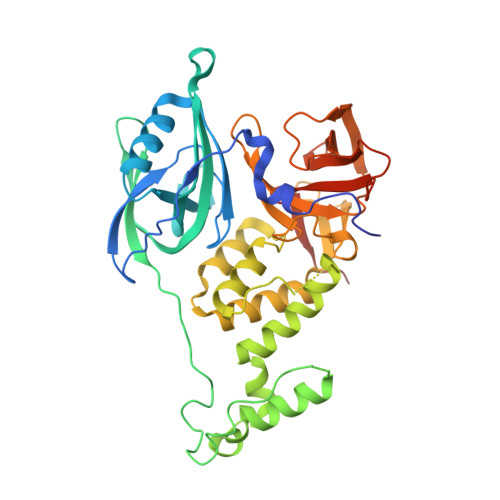
A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Bouguenina, H., Scarpino, A., O'Hanlon, J.A., Warne, J., Wang, H.Z., Wah Hak, L.C., Sadok, A., McAndrew, P.C., Stubbs, M., Pierrat, O.A., Hahner, T., Cabry, M.P., Le Bihan, Y.V., Mitsopoulos, C., Sialana, F.J., Roumeliotis, T.I., Burke, R., van Montfort, R.L.M., Choudhari, J., Chopra, R., Caldwell, J.J., Collins, I.(2023) Chembiochem 24: e202300351-e202300351
- PubMed: 37418539
- DOI: https://doi.org/10.1002/cbic.202300351
- Primary Citation of Related Structures:
8OIZ, 8OJH - PubMed Abstract:
Small molecules inducing protein degradation are important pharmacological tools to interrogate complex biology and are rapidly translating into clinical agents. However, to fully realise the potential of these molecules, selectivity remains a limiting challenge. Herein, we addressed the issue of selectivity in the design of CRL4 CRBN recruiting PROteolysis TArgeting Chimeras (PROTACs). Thalidomide derivatives used to generate CRL4 CRBN recruiting PROTACs have well described intrinsic monovalent degradation profiles by inducing the recruitment of neo-substrates, such as GSPT1, Ikaros and Aiolos. We leveraged structural insights from known CRL4 CRBN neo-substrates to attenuate and indeed remove this monovalent degradation function in well-known CRL4 CRBN molecular glues degraders, namely CC-885 and Pomalidomide. We then applied these design principles on a previously published BRD9 PROTAC (dBRD9-A) and generated an analogue with improved selectivity profile. Finally, we implemented a computational modelling pipeline to show that our degron blocking design does not impact PROTAC-induced ternary complex formation. We believe that the tools and principles presented in this work will be valuable to support the development of targeted protein degradation.
- Centre for Cancer Drug Discovery, Institute of Cancer Research, 15 Cotswold Road, Sutton, London, SM2 5NG, UK.
Organizational Affiliation: