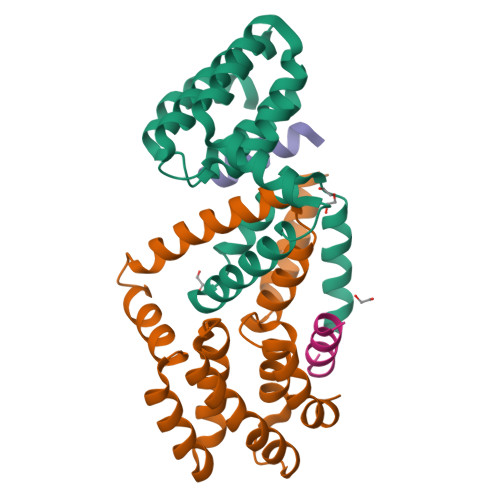
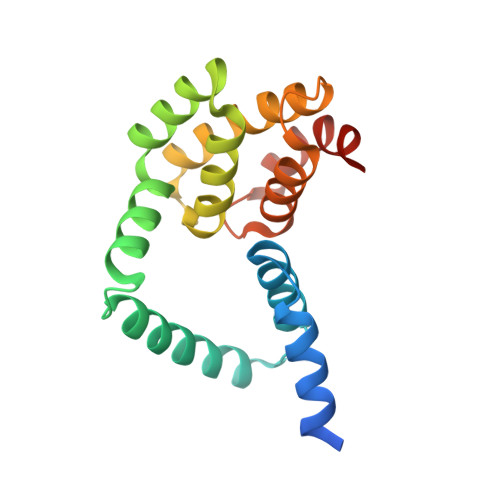
Structural basis for binding of Drosophila Smaug to the GPCR Smoothened and to the germline inducer Oskar.
Kubikova, J., Ubartaite, G., Metz, J., Jeske, M.(2023) Proc Natl Acad Sci U S A 120: e2304385120-e2304385120
- PubMed: 37523566
- DOI: https://doi.org/10.1073/pnas.2304385120
- Primary Citation of Related Structures:
8OIJ, 8OIK - PubMed Abstract:
Drosophila Smaug and its orthologs comprise a family of mRNA repressor proteins that exhibit various functions during animal development. Smaug proteins contain a characteristic RNA-binding sterile-α motif (SAM) domain and a conserved but uncharacterized N-terminal domain (NTD). Here, we resolved the crystal structure of the NTD of the human SAM domain-containing protein 4A (SAMD4A, a.k.a. Smaug1) to 1.6 Å resolution, which revealed its composition of a homodimerization D subdomain and a subdomain with similarity to a pseudo-HEAT-repeat analogous topology (PHAT) domain. Furthermore, we show that Drosophila Smaug directly interacts with the Drosophila germline inducer Oskar and with the Hedgehog signaling transducer Smoothened through its NTD. We determined the crystal structure of the NTD of Smaug in complex with a Smoothened α-helical peptide to 2.0 Å resolution. The peptide binds within a groove that is formed by both the D and PHAT subdomains. Structural modeling supported by experimental data suggested that an α-helix within the disordered region of Oskar binds to the NTD of Smaug in a mode similar to Smoothened. Together, our data uncover the NTD of Smaug as a peptide-binding domain.
Organizational Affiliation:
Biochemistry Center, Heidelberg University, Heidelberg 69120, Germany.