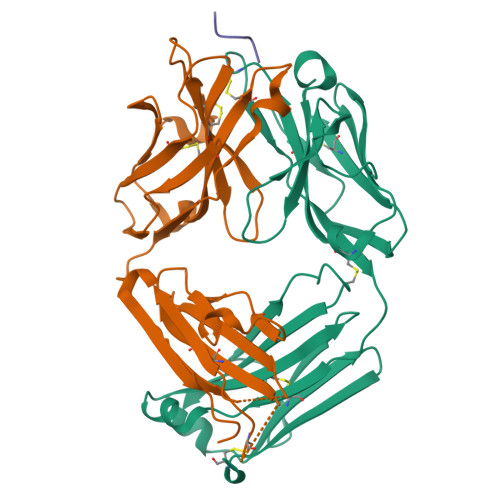
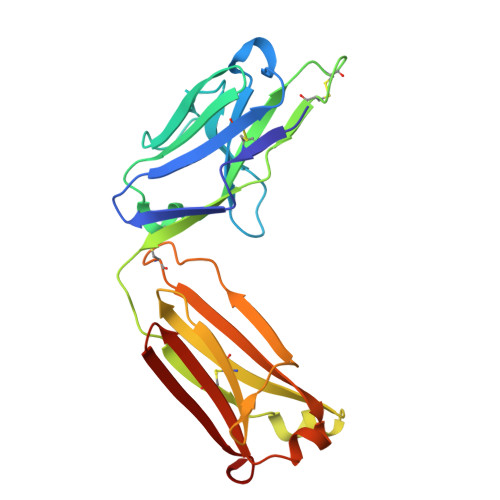
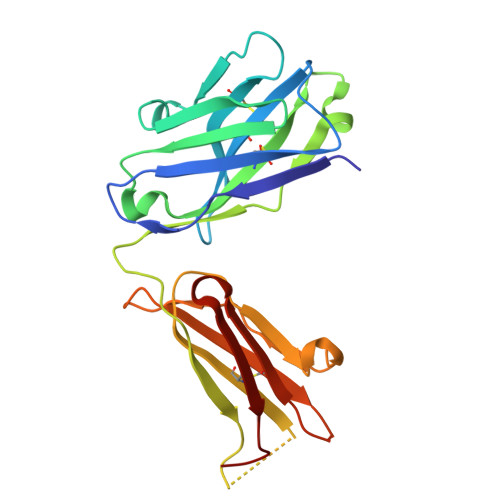
Structural basis of epitope recognition by anti-alpha-synuclein antibodies MJFR14-6-4-2.
Lieknina, I., Reimer, L., Pantelejevs, T., Lends, A., Jaudzems, K., El-Turabi, A., Gram, H., Hammi, A., Jensen, P.H., Tars, K.(2024) NPJ Parkinsons Dis 10: 206-206
- PubMed: 39463404
- DOI: https://doi.org/10.1038/s41531-024-00822-y
- Primary Citation of Related Structures:
8OG0 - PubMed Abstract:
Alpha-synuclein (α-syn) inclusions in the brain are hallmarks of so-called Lewy body diseases. Lewy bodies contain mainly aggregated α-syn together with some other proteins. Monomeric α-syn lacks a well-defined three-dimensional structure, but it can aggregate into oligomeric and fibrillar amyloid species, which can be detected using specific antibodies. Here we investigate the aggregate specificity of monoclonal MJFR14-6-4-2 antibodies. We conclude that partial masking of epitope in unstructured monomer in combination with a high local concentration of epitopes is the main reason for MJFR14-6-4-2 selectivity towards aggregates. Based on the structural insight, we produced mutant α-syn that when fibrillated is unable to bind MJFR14-6-4-2. Using these fibrils as a tool for seeding cellular α-syn aggregation, provides superior signal/noise ratio for detection of cellular α-syn aggregates by MJFR14-6-4-2. Our data provide a molecular level understanding of specific recognition of toxic amyloid oligomers, which is critical for the development of inhibitors against synucleinopathies.
Organizational Affiliation:
Latvian Biomedical Research and Study Centre, Ratsupites 1, k-1, LV-1067, Riga, Latvia.