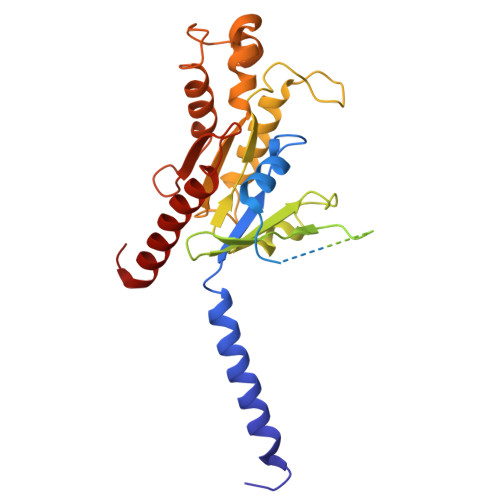
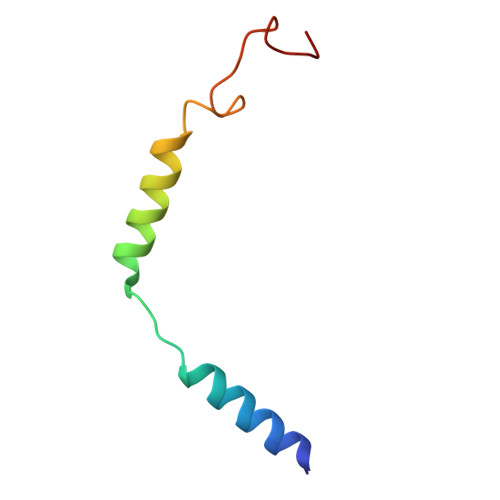
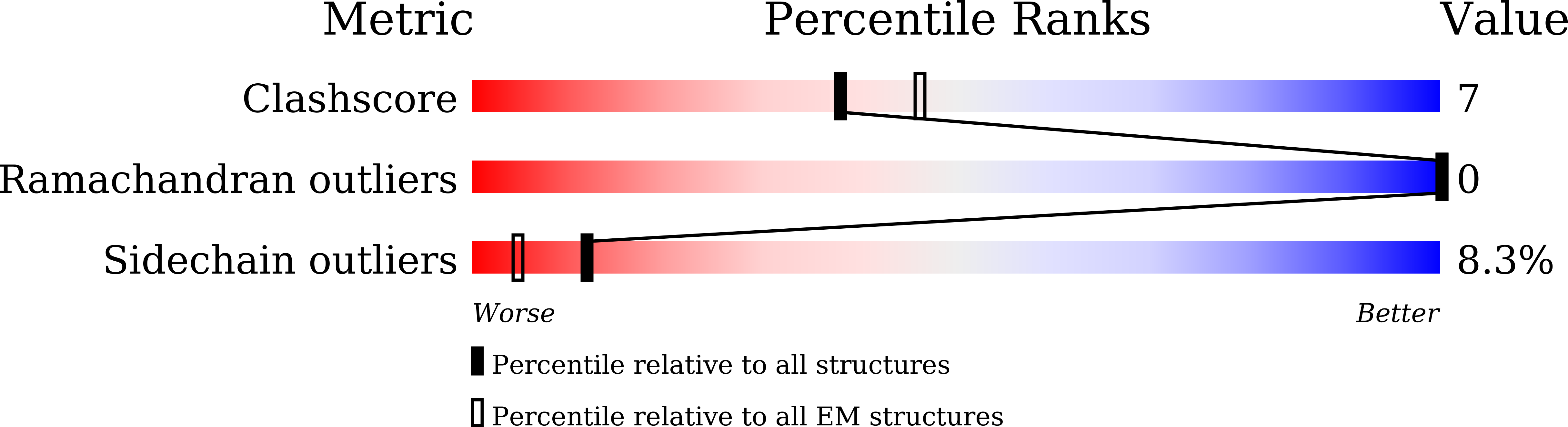Structural insights into KSHV-GPCR constitutive activation and CXCL1 chemokine recognition.
Liu, A., Liu, Y., Llinas Del Torrent Masachs, C., Zhang, W., Pardo, L., Ye, R.D.(2024) Proc Natl Acad Sci U S A 121: e2403217121-e2403217121
- PubMed: 39378089
- DOI: https://doi.org/10.1073/pnas.2403217121
- Primary Citation of Related Structures:
8K4O, 8K4P - PubMed Abstract:
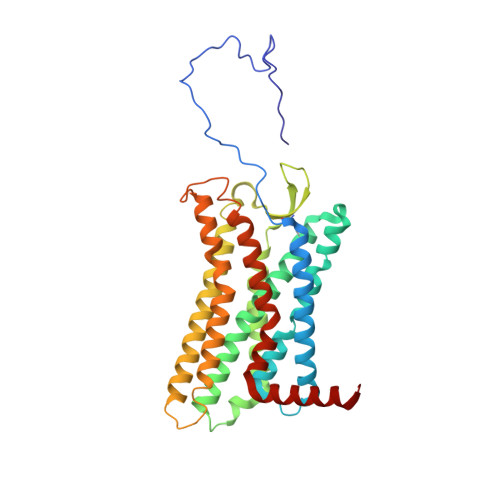
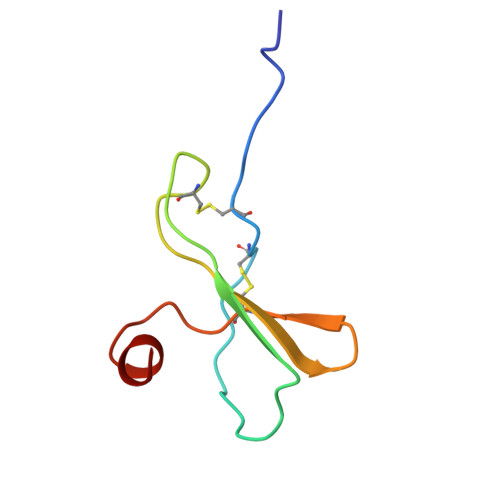
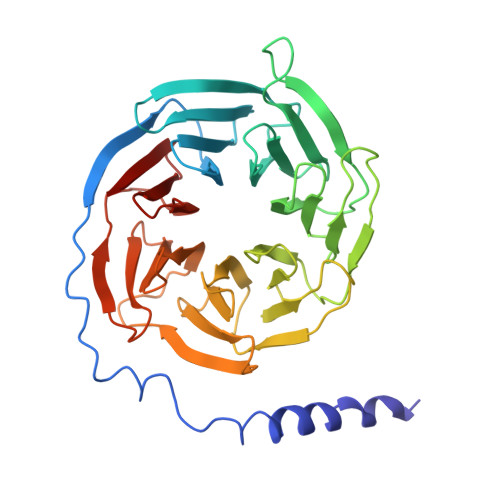
Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a viral G protein-coupled receptor, KSHV-GPCR, that contributes to KSHV immune evasion and pathogenesis of Kaposi's sarcoma. KSHV-GPCR shares a high similarity with CXC chemokine receptors CXCR2 and can be activated by selected chemokine ligands. Like other herpesvirus-encoded GPCRs, KSHV-GPCR is characterized by its constitutive activity by coupling to various G proteins. We investigated the structural basis of ligand-dependent and constitutive activation of KSHV-GPCR, obtaining high-resolution cryo-EM structures of KSHV-GPCR-Gi complexes with and without the bound CXCL1 chemokine. Analysis of the apo-KSHV-GPCR-Gi structure (2.81 Å) unraveled the involvement of extracellular loop 2 in constitutive activation of the receptor. In comparison, the CXCL1-bound KSHV-GPCR-Gi structure (3.01 Å) showed a two-site binding mode and provided detailed information of CXCL1 binding to a chemokine receptor. The dual activation mechanism employed by KSHV-GPCR represents an evolutionary adaptation for immune evasion and contributes to the pathogenesis of Kaposi's sarcoma. Together with results from functional assays that confirmed the structural models, these findings may help to develop therapeutic strategies for KSHV infection.
- Kobilka Institute of Innovative Drug Discovery, School of Medicine, The Chinese University of Hong Kong, Shenzhen, Guangdong 518172, China.
Organizational Affiliation: