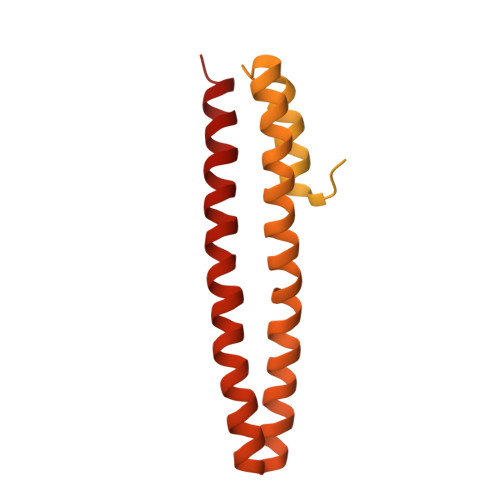
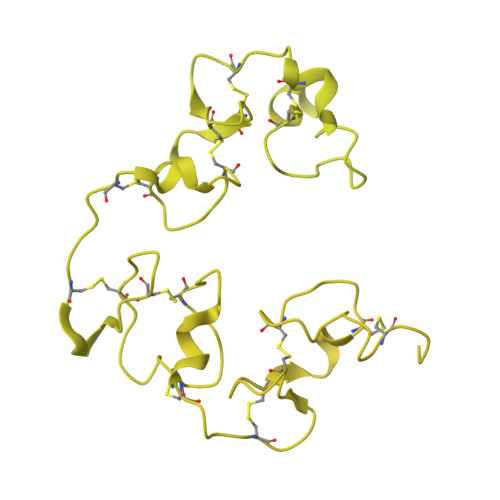
Cryo-EM structures elucidate the multiligand receptor nature of megalin.
Goto, S., Tsutsumi, A., Lee, Y., Hosojima, M., Kabasawa, H., Komochi, K., Nagatoishi, S., Takemoto, K., Tsumoto, K., Nishizawa, T., Kikkawa, M., Saito, A.(2024) Proc Natl Acad Sci U S A 121: e2318859121-e2318859121
- PubMed: 38771880
- DOI: https://doi.org/10.1073/pnas.2318859121
- Primary Citation of Related Structures:
8JUT, 8JUU, 8JX8, 8JX9, 8JXA, 8JXB, 8JXC, 8JXD, 8JXE, 8JXF, 8JXG, 8JXH, 8JXI, 8JXJ - PubMed Abstract:
Megalin (low-density lipoprotein receptor-related protein 2) is a giant glycoprotein of about 600 kDa, mediating the endocytosis of more than 60 ligands, including those of proteins, peptides, and drug compounds [S. Goto, M. Hosojima, H. Kabasawa, A. Saito, Int. J. Biochem. Cell Biol. 157 , 106393 (2023)]. It is expressed predominantly in renal proximal tubule epithelial cells, as well as in the brain, lungs, eyes, inner ear, thyroid gland, and placenta. Megalin is also known to mediate the endocytosis of toxic compounds, particularly those that cause renal and hearing disorders [Y. Hori et al. , J. Am. Soc. Nephrol. 28 , 1783-1791 (2017)]. Genetic megalin deficiency causes Donnai-Barrow syndrome/facio-oculo-acoustico-renal syndrome in humans. However, it is not known how megalin interacts with such a wide variety of ligands and plays pathological roles in various organs. In this study, we elucidated the dimeric architecture of megalin, purified from rat kidneys, using cryoelectron microscopy. The maps revealed the densities of endogenous ligands bound to various regions throughout the dimer, elucidating the multiligand receptor nature of megalin. We also determined the structure of megalin in complex with receptor-associated protein, a molecular chaperone for megalin. The results will facilitate further studies on the pathophysiology of megalin-dependent multiligand endocytic pathways in multiple organs and will also be useful for the development of megalin-targeted drugs for renal and hearing disorders, Alzheimer's disease [B. V. Zlokovic et al. , Proc. Natl. Acad. Sci. U.S.A. 93 , 4229-4234 (1996)], and other illnesses.
- Department of Applied Molecular Medicine, Kidney Research Center, Niigata University Graduate School of Medical and Dental Sciences, Niigata City 951-8510, Japan.
Organizational Affiliation: