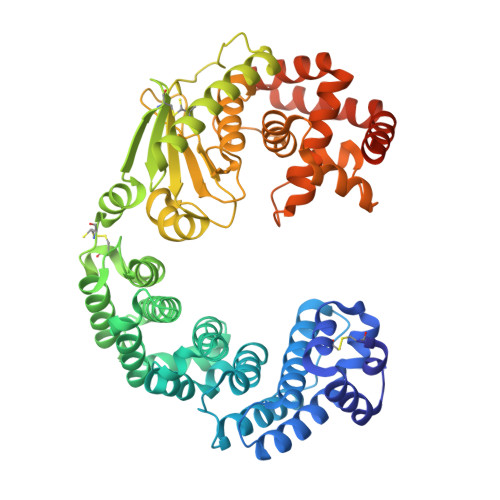
Insights into the catalytic mechanism of Grimontia hollisae collagenase through structural and mutational analyses.
Ueshima, S., Yasumoto, M., Kitagawa, Y., Akazawa, K., Takita, T., Tanaka, K., Hattori, S., Mizutani, K., Mikami, B., Yasukawa, K.(2023) FEBS Lett 597: 2473-2483
- PubMed: 37698340
- DOI: https://doi.org/10.1002/1873-3468.14732
- Primary Citation of Related Structures:
8JT1 - PubMed Abstract:
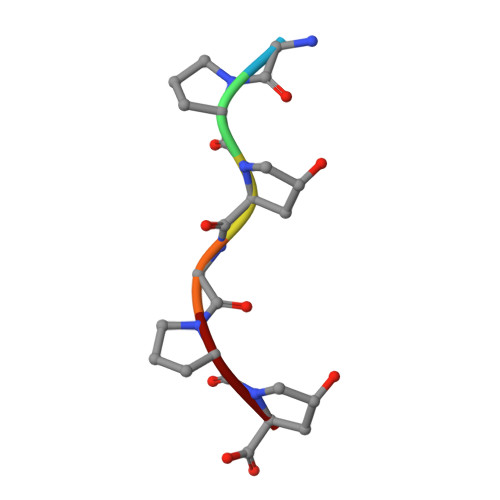
Grimontia hollisae collagenase (Ghcol) exhibits high collagen-degrading activity. To explore its catalytic mechanism, its substrate (Gly-Pro-Hyp-Gly-Pro-Hyp, GPOGPO)-complexed crystal structure was determined at 2.0 Å resolution. A water molecule was observed near the active-site zinc ion. Since this water was not observed in the product (GPO)-complexed Ghcol, it was hypothesized that the GPOGPO-complexed Ghcol structure reflects a Michaelis complex, providing a structural basis for understanding the catalytic mechanism. Analyses of the active-site geometry and site-directed mutagenesis of the active-site tyrosine residues revealed that Glu493 and Tyr564 were essential for catalysis, suggesting that Glu493 functions as an acid and base catalyst while Tyr564 stabilizes the tetrahedral complex in the transition state. These results shed light on the catalytic mechanism of bacterial collagenase.
- Division of Food Science and Biotechnology, Graduate School of Agriculture, Kyoto University, Japan.
Organizational Affiliation: