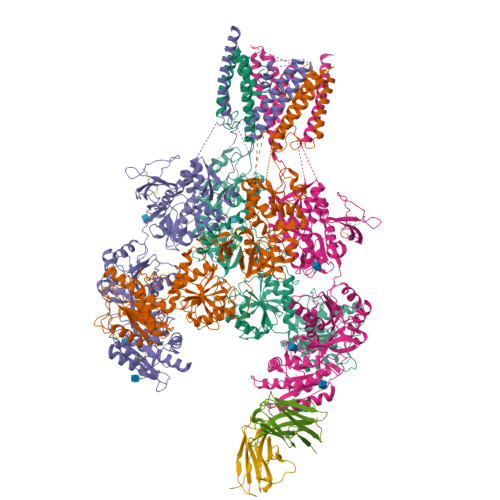
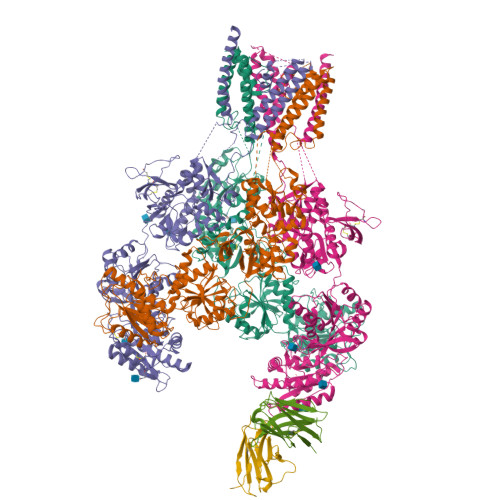
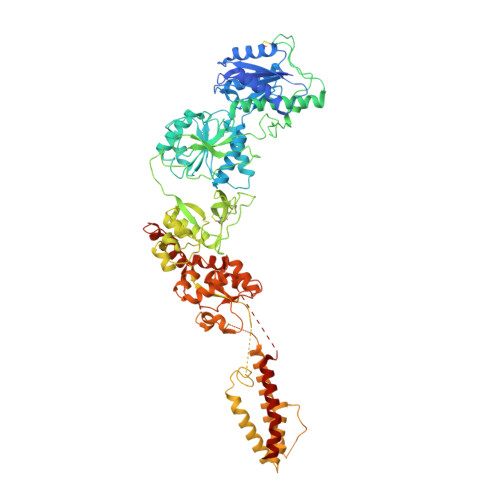
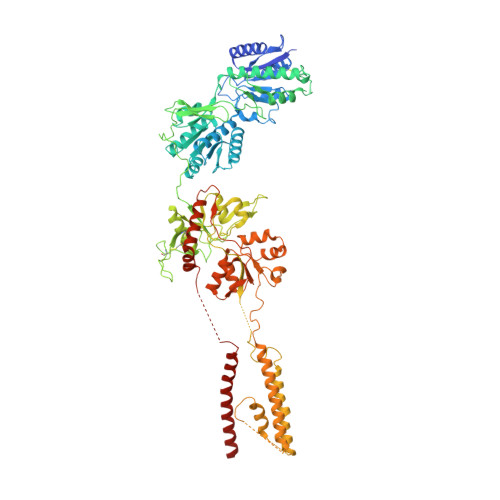
Structural basis for antibody-mediated NMDA receptor clustering and endocytosis in autoimmune encephalitis.
Wang, H., Xie, C., Deng, B., Ding, J., Li, N., Kou, Z., Jin, M., He, J., Wang, Q., Wen, H., Zhang, J., Zhou, Q., Chen, S., Chen, X., Yuan, T.F., Zhu, S.(2024) Nat Struct Mol Biol 31: 1987-1996
- PubMed: 39227720
- DOI: https://doi.org/10.1038/s41594-024-01387-3
- Primary Citation of Related Structures:
8JIZ, 8JJ0, 8JJ1, 8JJ2 - PubMed Abstract:
Antibodies against N-methyl-D-aspartate receptors (NMDARs) are most frequently detected in persons with autoimmune encephalitis (AE) and used as diagnostic biomarkers. Elucidating the structural basis of monoclonal antibody (mAb) binding to NMDARs would facilitate the development of targeted therapy for AE. Here, we reconstructed nanodiscs containing green fluorescent protein-fused NMDARs to label and sort individual immune B cells from persons with AE and further cloned and identified mAbs against NMDARs. This allowed cryo-electron microscopy analysis of NMDAR-Fab complexes, revealing that autoantibodies bind to the R1 lobe of the N-terminal domain of the GluN1 subunit. Small-angle X-ray scattering studies demonstrated NMDAR-mAb stoichiometry of 2:1 or 1:2, structurally suitable for mAb-induced clustering and endocytosis of NMDARs. Importantly, these mAbs reduced the surface NMDARs and NMDAR-mediated currents, without tonically affecting NMDAR channel gating. These structural and functional findings imply that the design of neutralizing antibody binding to the R1 lobe of NMDARs represents a potential therapy for AE treatment.
Organizational Affiliation:
Institute of Neuroscience, CAS Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, Shanghai, China.