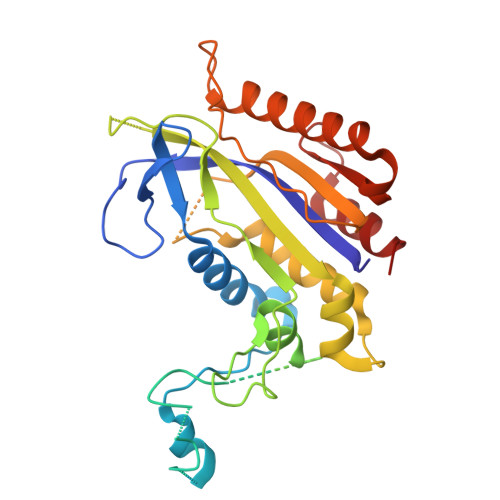
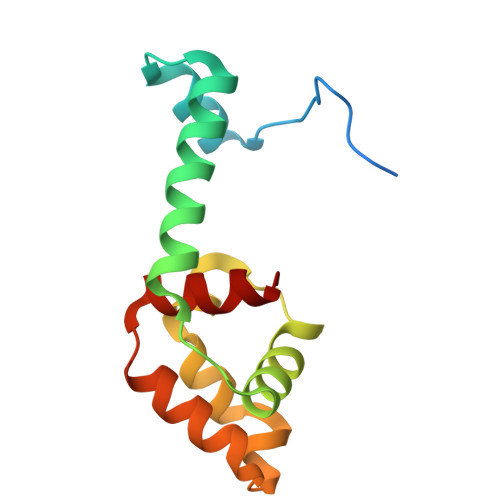
A new anti-CRISPR gene promotes the spread of drug-resistance plasmids in Klebsiella pneumoniae.
Jiang, C., Yu, C., Sun, S., Lin, J., Cai, M., Wei, Z., Feng, L., Li, J., Zhang, Y., Dong, K., Guo, X., Qin, J., Zhang, Y.(2024) Nucleic Acids Res 52: 8370-8384
- PubMed: 38888121
- DOI: https://doi.org/10.1093/nar/gkae516
- Primary Citation of Related Structures:
8JI9 - PubMed Abstract:
The Klebsiella pneumoniae (K. pneumoniae, Kp) populations carrying both resistance-encoding and virulence-encoding mobile genetic elements (MGEs) significantly threaten global health. In this study, we identified a new anti-CRISPR gene (acrIE10) on a conjugative plasmid with self-target sequence in K. pneumoniae with type I-E* CRISPR-Cas system. AcrIE10 interacts with the Cas7* subunit of K. pneumoniae I-E* CRISPR-Cas system. The crystal structure of the AcrIE10-KpCas7* complex suggests that AcrIE10 suppresses the I-E* CRISPR-Cas by binding directly to Cas7 to prevent its hexamerization, thereby preventing the surveillance complex assembly and crRNA loading. Bioinformatic and functional analyses revealed that AcrIE10 is functionally widespread across diverse species. Our study reports a novel anti-CRISPR and highlights its potential role in spreading resistance and virulence among pathogens.
Organizational Affiliation:
Department of Microbiology and Immunology, Shanghai Jiao Tong University School of Medicine, Shanghai 200025, China.