Mechanistic Insights into the Interactions of Arl8b with the RUN Domains of PLEKHM1 and SKIP.
Qiu, X., Li, Y., Wang, Y., Gong, X., Wang, Y., Pan, L.(2023) J Mol Biol 435: 168293-168293
- PubMed: 37775038
- DOI: https://doi.org/10.1016/j.jmb.2023.168293
- Primary Citation of Related Structures:
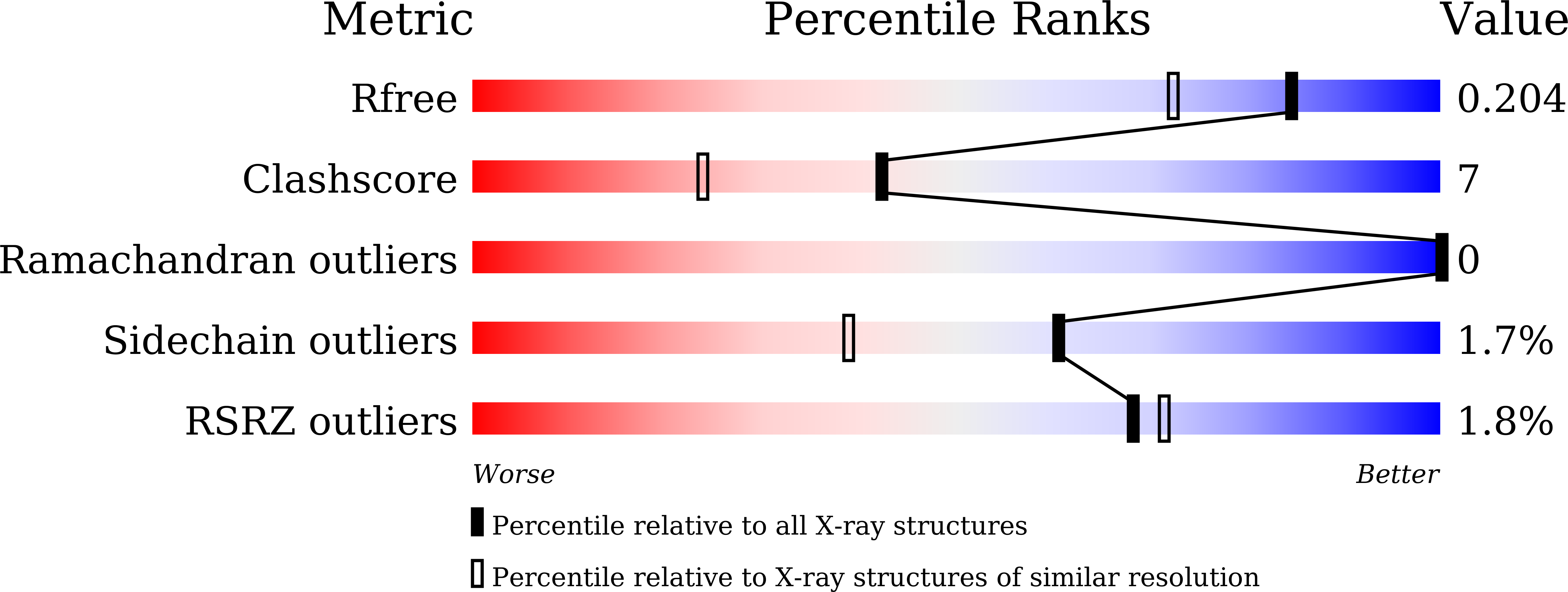
8JC5, 8JCA - PubMed Abstract:
Arl8b, a specific Arf-like family GTPase present on lysosome, and plays critical roles in many lysosome-related cellular processes such as autophagy. The active Arl8b can be specifically recognized by the RUN domains of two Arl8b-effectors PLEKHM1 and SKIP, thereby regulating the autophagosome/lysosome membrane fusion and the intracellular lysosome positioning, respectively. However, the mechanistic bases underlying the interactions of Arl8b with the RUN domains of PLEKHM1 and SKIP remain elusive. Here, we report the two high-resolution crystal structures of the active Arl8b in complex with the RUN domains of PLEKHM1 and SKIP. In addition to elucidating the detailed molecular mechanism governing the specific interactions of the active Arl8b with the RUN domains of PLEKHM1 and SKIP, the determined complex structures also reveal a general binding mode shared by the PLEKHM1 and SKIP RUN domains for interacting with the active Arl8b. Furthermore, we uncovered a competitive relationship between the RUN domains of PLEKHM1 and SKIP in binding to the active Arl8b as well as a unique small GTPase-binding mode adopted by the PLEKHM1 and SKIP RUN domains, thereby enriching the repertoire of the RUN domain/small GTPase interaction modes. In all, our findings provide new mechanistic insights into the interactions of the active Arl8b with PLEKHM1 and SKIP, and are valuable for further understanding the working modes of these proteins in relevant cellular processes.
Organizational Affiliation:
College of Chemistry and Materials Science, Sichuan Normal University, Chengdu, Sichuan 610068, China; State Key Laboratory of Chemical Biology, Shanghai Institute of Organic Chemistry, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai 200032, China.