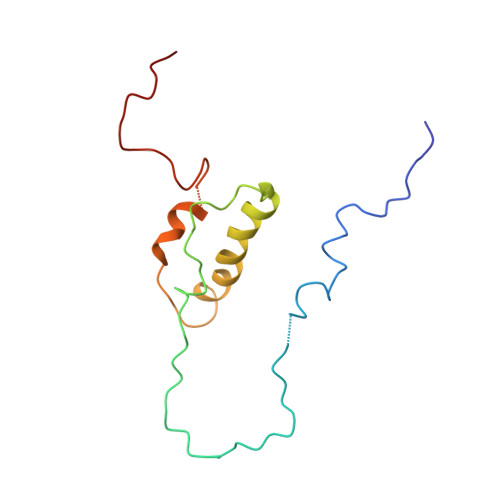
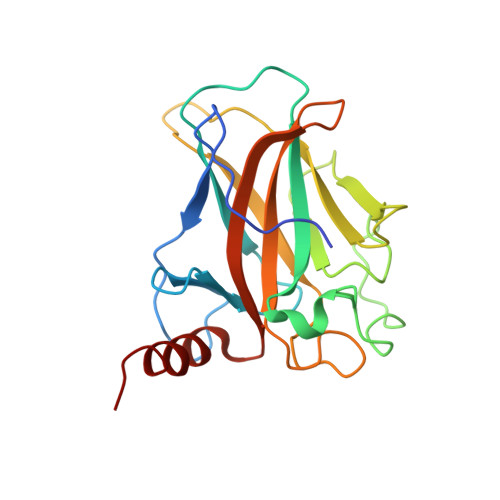
Influence of the interaction between p53 and ZNF568 on mitochondrial oxidative phosphorylation.
Han, C.W., Jeong, M.S., Jang, S.B.(2024) Int J Biol Macromol 275: 133314-133314
- PubMed: 38944084
- DOI: https://doi.org/10.1016/j.ijbiomac.2024.133314
- Primary Citation of Related Structures:
8J8N - PubMed Abstract:
The tumor suppressor p53 plays important roles in suppressing the development and progression of cancer by responding to various stress signals. In addition, p53 can regulate the metabolic pathways of cancer cells by regulating energy metabolism and oxidative phosphorylation. Here, we present a mechanism for the interaction between p53 and ZNF568. Initially, we used X-ray crystallography to determine the irregular loop structure of the ZNF568 KRAB domain; this loop plays an important role in the interaction between p53 and ZNF568. In addition, Cryo-EM was used to examine how the p53 DBD and ZNF568 KRAB domains bind together. The function of ZNF568 on p53-mediated mitochondrial respiration was confirmed by measuring glucose consumption and lactate production. These findings show that ZNF568 can reduce p53-mediated mitochondrial respiratory activity by binding to p53 and inhibiting the transcription of SCO2. SIGNIFICANCE: ZNF568 can directly bind to the p53 DBD and transcriptionally regulate the SCO2 gene. SCO2 transcriptional regulation by interaction between ZNF568 and p53 may regulate the balance between mitochondrial respiration and glycolysis.
- Institute of Systems Biology, Pusan National University, Jangjeon-dong, Geumjeong-gu, Busan 46241, Republic of Korea.
Organizational Affiliation: