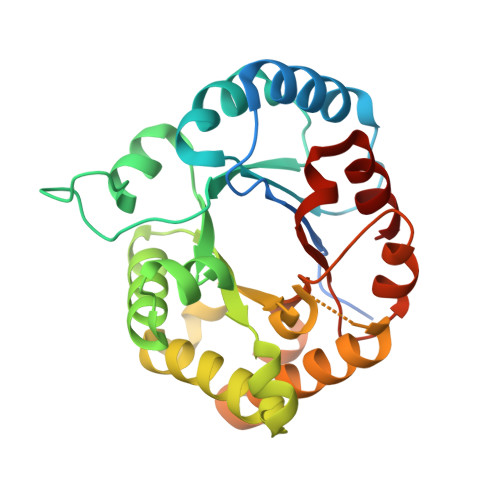
Leishmania orientalis triosephosphate isomerase crystal structure at 1.45 angstroms resolution and its potential specific inhibitors
Kuaprasert, B., Leartsakulpanich, U., Riangrungroj, P., Pornthanakasem, W., Suginta, W., Robinson, R., Zhou, Y., Mungthin, M., Leelayoova, S., Saehlee, S., Kiriwan, D., Choowongkomon, K.To be published.