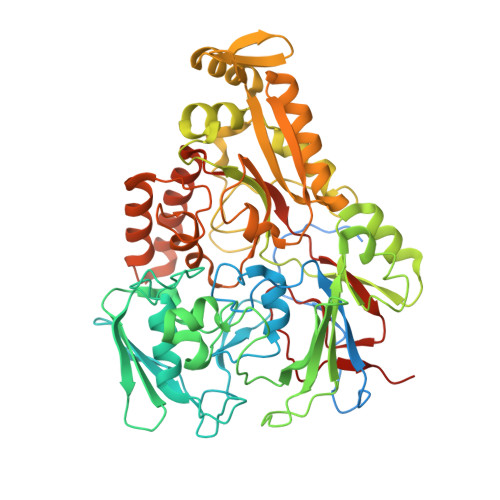
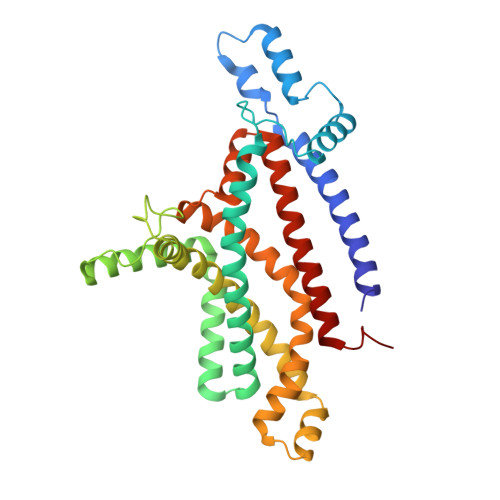
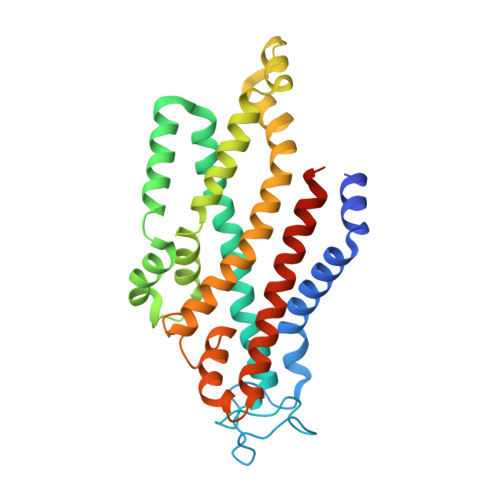
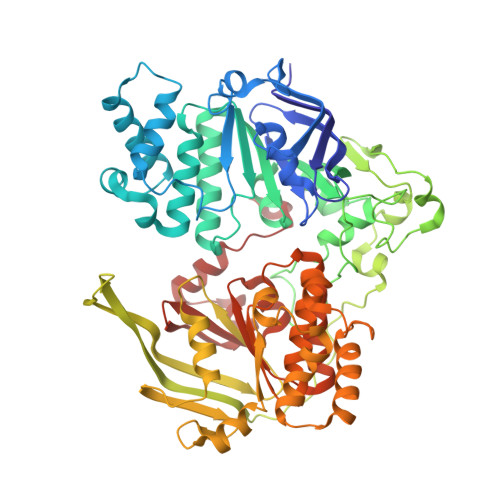
An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Yang, X., Hu, T., Liang, J., Xiong, Z., Lin, Z., Zhao, Y., Zhou, X., Gao, Y., Sun, S., Yang, X., Guddat, L.W., Yang, H., Rao, Z., Zhang, B.(2024) Nat Struct Mol Biol 31: 1072-1082
- PubMed: 38548954
- DOI: https://doi.org/10.1038/s41594-024-01256-z
- Primary Citation of Related Structures:
8J5Q, 8J5R, 8J5S, 8J5T, 8J5U - PubMed Abstract:
Oligopeptide permease, OppABCD, belongs to the type I ABC transporter family. Its role is to import oligopeptides into bacteria for nutrient uptake and to modulate the host immune response. OppABCD consists of a cluster C substrate-binding protein (SBP), OppA, membrane-spanning OppB and OppC subunits, and an ATPase, OppD, that contains two nucleotide-binding domains (NBDs). Here, using cryo-electron microscopy, we determined the high-resolution structures of Mycobacterium tuberculosis OppABCD in the resting state, oligopeptide-bound pre-translocation state, AMPPNP-bound pre-catalytic intermediate state and ATP-bound catalytic intermediate state. The structures show an assembly of a cluster C SBP with its ABC translocator and a functionally required [4Fe-4S] cluster-binding domain in OppD. Moreover, the ATP-bound OppABCD structure has an outward-occluded conformation, although no substrate was observed in the transmembrane cavity. Here, we reveal an oligopeptide recognition and translocation mechanism of OppABCD, which provides a perspective on how this and other type I ABC importers facilitate bulk substrate transfer across the lipid bilayer.
- Shanghai Institute for Advanced Immunochemical Studies and School of Life Science and Technology, ShanghaiTech University, Shanghai, China. yangxl@shanghaitech.edu.cn.
Organizational Affiliation: