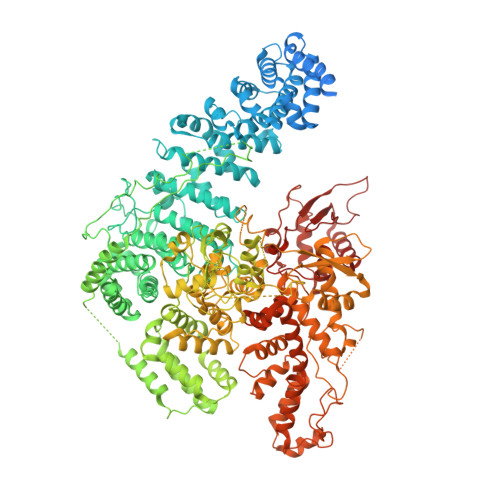
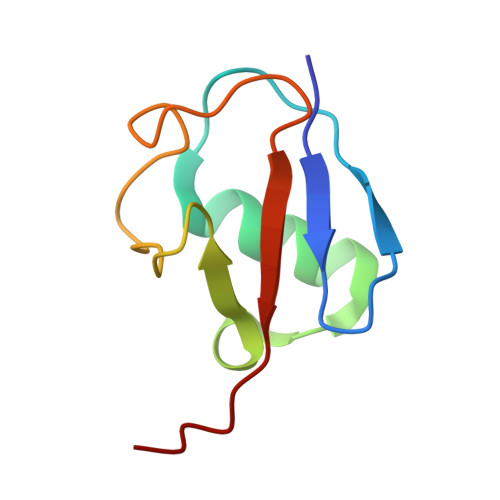
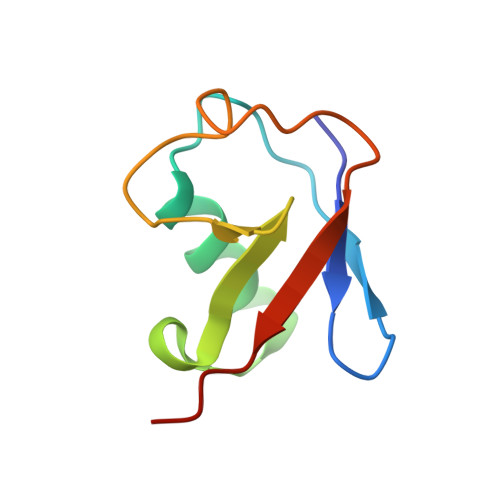
Structural visualization of HECT-type E3 ligase Ufd4 accepting and transferring ubiquitin to form K29/K48-branched polyubiquitination.
Wu, X., Ai, H., Mao, J., Cai, H., Liang, L.J., Tong, Z., Deng, Z., Zheng, Q., Liu, L., Pan, M.(2025) Nat Commun 16: 4313-4313
- PubMed: 40341121
- DOI: https://doi.org/10.1038/s41467-025-59569-6
- Primary Citation of Related Structures:
8J1P, 8J1R, 9KEN - PubMed Abstract:
The K29/K48-linked ubiquitination generated by the cooperative catalysis of E3 ligase Ufd4 and Ubr1 is an enhanced protein degradation signal, in which Ufd4 is responsible for introducing K29-linked ubiquitination to K48-linked ubiquitin chains to augment polyubiquitination. How HECT-E3 ligase Ufd4 mediates the ubiquitination event remains unclear. Here, we biochemically determine that Ufd4 preferentially catalyses K29-linked ubiquitination on K48-linked ubiquitin chains to generate K29/K48-branched ubiquitin chains and capture structural snapshots of Ub transfer cascades for Ufd4-mediated ubiquitination. The N-terminal ARM region and HECT domain C-lobe of Ufd4 are identified and characterized as key structural elements that together recruit K48-linked diUb and orient Lys29 of its proximal Ub to the active cysteine of Ufd4 for K29-linked branched ubiquitination. These structures not only provide mechanistic insights into the architecture of the Ufd4 complex but also provide structural visualization of branched ubiquitin chain formation by a HECT-type E3 ligase.
- Institute of Translational Medicine, School of Pharmaceutical Sciences, School of Chemistry and Chemical Engineering, National Center for Translational Medicine (Shanghai), Shanghai Key Laboratory for Antibody-Drug Conjugates with Innovative Target, Shanghai Jiao Tong University, Shanghai, China.
Organizational Affiliation: