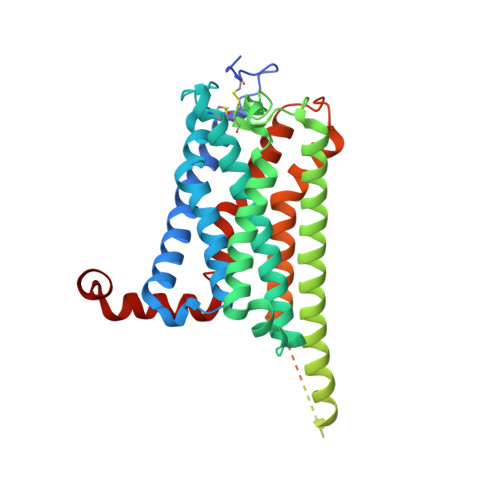
Structural insights into ligand recognition and activation of the medium-chain fatty acid-sensing receptor GPR84.
Liu, H., Zhang, Q., He, X., Jiang, M., Wang, S., Yan, X., Cheng, X., Liu, Y., Nan, F.J., Xu, H.E., Xie, X., Yin, W.(2023) Nat Commun 14: 3271-3271
- PubMed: 37277332
- DOI: https://doi.org/10.1038/s41467-023-38985-6
- Primary Citation of Related Structures:
8J18, 8J19, 8J1A - PubMed Abstract:
GPR84 is an orphan class A G protein-coupled receptor (GPCR) that is predominantly expressed in immune cells and plays important roles in inflammation, fibrosis, and metabolism. Here, we present cryo-electron microscopy (cryo-EM) structures of Gα i protein-coupled human GPR84 bound to a synthetic lipid-mimetic ligand, LY237, or a putative endogenous ligand, a medium-chain fatty acid (MCFA) 3-hydroxy lauric acid (3-OH-C12). Analysis of these two ligand-bound structures reveals a unique hydrophobic nonane tail -contacting patch, which forms a blocking wall to select MCFA-like agonists with the correct length. We also identify the structural features in GPR84 that coordinate the polar ends of LY237 and 3-OH-C12, including the interactions with the positively charged side chain of R172 and the downward movement of the extracellular loop 2 (ECL2). Together with molecular dynamics simulations and functional data, our structures reveal that ECL2 not only contributes to direct ligand binding, but also plays a pivotal role in ligand entry from the extracellular milieu. These insights into the structure and function of GPR84 could improve our understanding of ligand recognition, receptor activation, and Gα i -coupling of GPR84. Our structures could also facilitate rational drug discovery against inflammation and metabolic disorders targeting GPR84.
- State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: