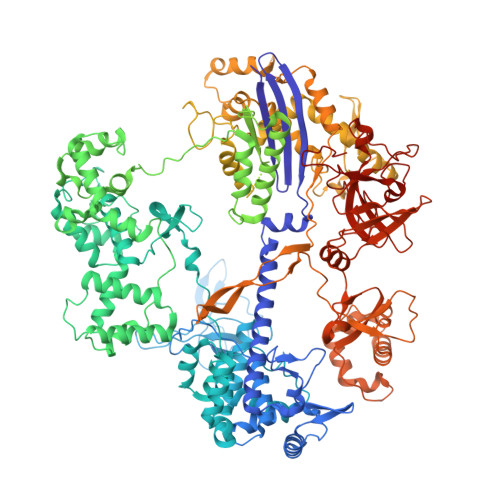
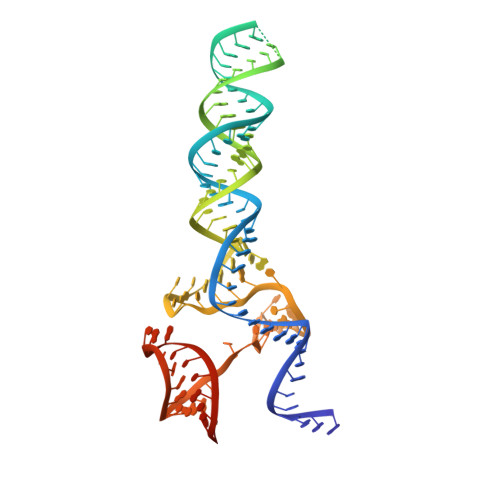
Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Zhang, S., Sun, A., Qian, J.M., Lin, S., Xing, W., Yang, Y., Zhu, H.Z., Zhou, X.Y., Guo, Y.S., Liu, Y., Meng, Y., Jin, S.L., Song, W., Li, C.P., Li, Z., Jin, S., Wang, J.H., Dong, M.Q., Gao, C., Chen, C., Bai, Y., Liu, J.G.(2024) Nature 630: 484-492
- PubMed: 38811729
- DOI: https://doi.org/10.1038/s41586-024-07486-x
- Primary Citation of Related Structures:
8IYQ, 8WMH, 8WMM, 8WMN, 8WR4 - PubMed Abstract:
The CRISPR system is an adaptive immune system found in prokaryotes that defends host cells against the invasion of foreign DNA 1 . As part of the ongoing struggle between phages and the bacterial immune system, the CRISPR system has evolved into various types, each with distinct functionalities 2 . Type II Cas9 is the most extensively studied of these systems and has diverse subtypes. It remains uncertain whether members of this family can evolve additional mechanisms to counter viral invasions 3,4 . Here we identify 2,062 complete Cas9 loci, predict the structures of their associated proteins and reveal three structural growth trajectories for type II-C Cas9. We found that novel associated genes (NAGs) tended to be present within the loci of larger II-C Cas9s. Further investigation revealed that CbCas9 from Chryseobacterium species contains a novel β-REC2 domain, and forms a heterotetrameric complex with an NAG-encoded CRISPR-Cas-system-promoting (pro-CRISPR) protein of II-C Cas9 (PcrIIC1). The CbCas9-PcrIIC1 complex exhibits enhanced DNA binding and cleavage activity, broader compatibility for protospacer adjacent motif sequences, increased tolerance for mismatches and improved anti-phage immunity, compared with stand-alone CbCas9. Overall, our work sheds light on the diversity and 'growth evolutionary' trajectories of II-C Cas9 proteins at the structural level, and identifies many NAGs-such as PcrIIC1, which serves as a pro-CRISPR factor to enhance CRISPR-mediated immunity.
- Beijing Frontier Research Center for Biological Structure, Beijing Advanced Innovation Center for Structural Biology, State Key Laboratory of Membrane Biology, Tsinghua-Peking Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing, China.
Organizational Affiliation: