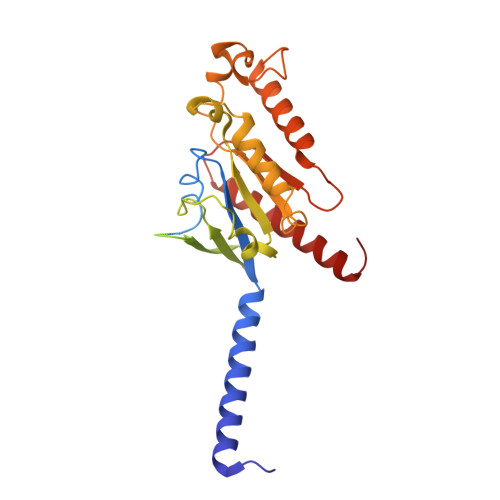
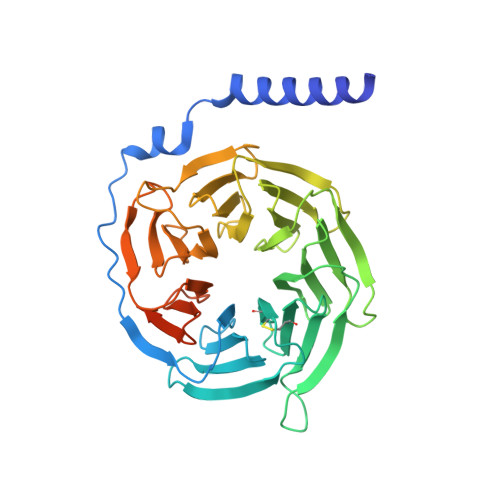
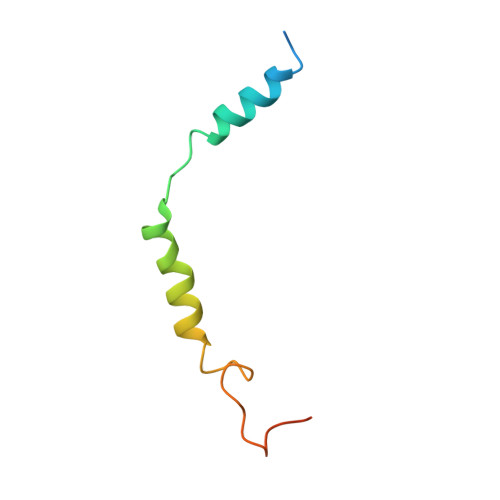
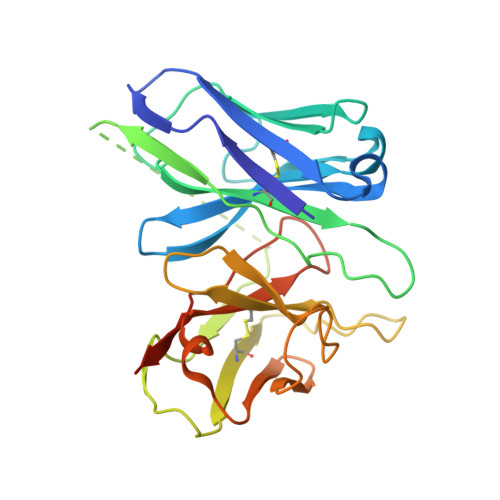
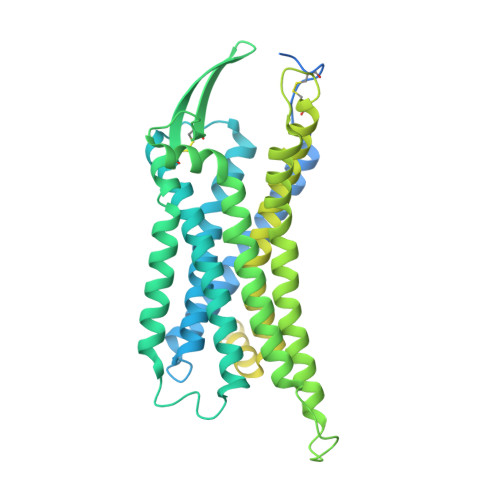
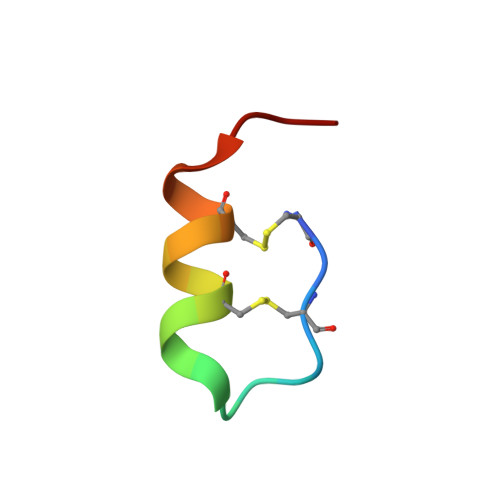
Cryo-EM structure of the endothelin-1-ET B -G i complex.
Sano, F.K., Akasaka, H., Shihoya, W., Nureki, O.(2023) Elife 12
- PubMed: 37096326
- DOI: https://doi.org/10.7554/eLife.85821
- Primary Citation of Related Structures:
8IY5, 8IY6 - PubMed Abstract:
The endothelin ET B receptor is a promiscuous G-protein coupled receptor that is activated by vasoactive peptide endothelins. ET B signaling induces reactive astrocytes in the brain and vasorelaxation in vascular smooth muscle. Consequently, ET B agonists are expected to be drugs for neuroprotection and improved anti-tumor drug delivery. Here, we report the cryo-electron microscopy structure of the endothelin-1-ET B -G i complex at 2.8 Å resolution, with complex assembly stabilized by a newly established method. Comparisons with the inactive ET B receptor structures revealed how endothelin-1 activates the ET B receptor. The NPxxY motif, essential for G-protein activation, is not conserved in ET B , resulting in a unique structural change upon G-protein activation. Compared with other GPCR-G-protein complexes, ET B binds G i in the shallowest position, further expanding the diversity of G-protein binding modes. This structural information will facilitate the elucidation of G-protein activation and the rational design of ET B agonists.
- Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Tokyo, Japan.
Organizational Affiliation: