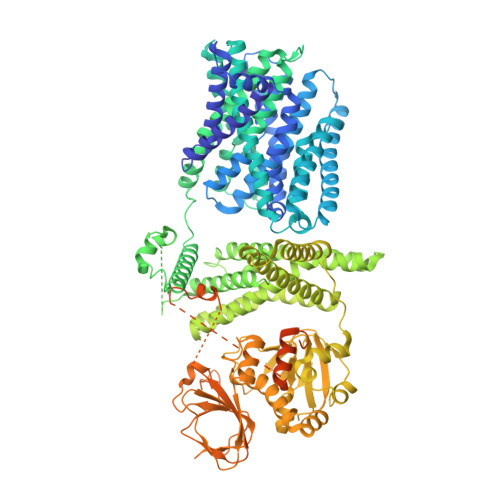
Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na + /H + antiporter.
Zhang, X.Y., Tang, L.H., Nie, J.W., Zhang, C.R., Han, X., Li, Q.Y., Qin, L., Wang, M.H., Huang, X., Yu, F., Su, M., Wang, Y., Xu, R.M., Guo, Y., Xie, Q., Chen, Y.H.(2023) Nat Plants 9: 1924-1936
- PubMed: 37884653
- DOI: https://doi.org/10.1038/s41477-023-01551-5
- Primary Citation of Related Structures:
8IWO, 8J2M - PubMed Abstract:
Salinity is one of the most severe abiotic stresses that adversely affect plant growth and agricultural productivity. The plant Na + /H + antiporter Salt Overly Sensitive 1 (SOS1) located in the plasma membrane extrudes excess Na + out of cells in response to salt stress and confers salt tolerance. However, the molecular mechanism underlying SOS1 activation remains largely elusive. Here we elucidate two cryo-electron microscopy structures of rice (Oryza sativa) SOS1, a full-length protein in an auto-inhibited state and a truncated version in an active state. The SOS1 forms a dimeric architecture, with an NhaA-folded transmembrane domain portion in the membrane and an elongated cytosolic portion of multiple regulatory domains in the cytoplasm. The structural comparison shows that SOS1 adopts an elevator transport mechanism accompanied by a conformational transition of the highly conserved Pro 148 in the unwound transmembrane helix 5 (TM 5 ), switching from an occluded conformation in the auto-inhibited state to a conducting conformation in the active state. These findings allow us to propose an inhibition-release mechanism for SOS1 activation and elucidate how SOS1 controls Na + homeostasis in response to salt stress.
- State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: