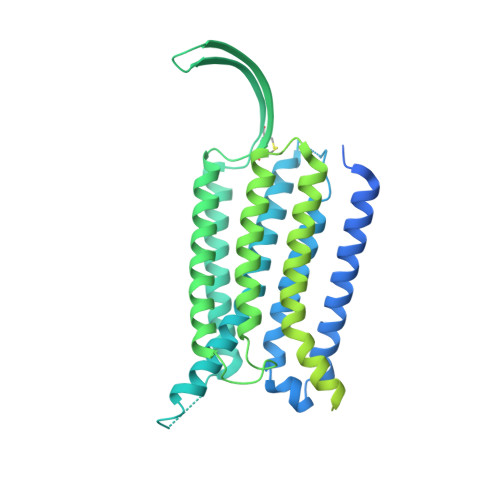
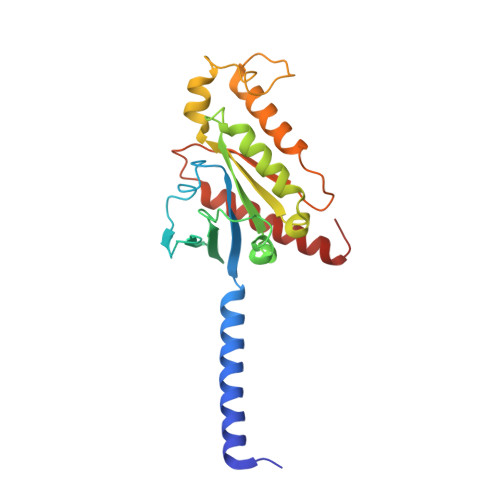
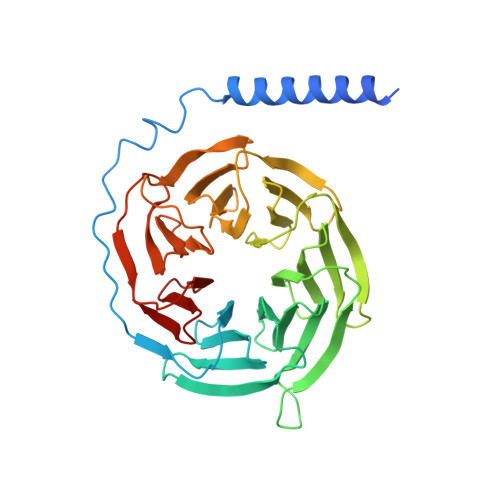
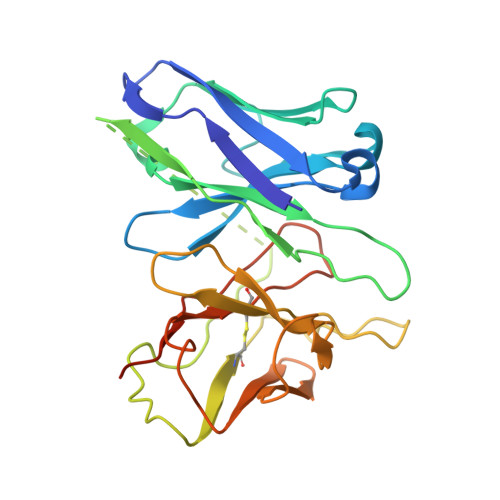
Constitutive activation mechanism of a class C GPCR.
Shin, J., Park, J., Jeong, J., Lam, J.H., Qiu, X., Wu, D., Kim, K., Lee, J.Y., Robinson, C.V., Hyun, J., Katritch, V., Kim, K.P., Cho, Y.(2024) Nat Struct Mol Biol 31: 678-687
- PubMed: 38332368
- DOI: https://doi.org/10.1038/s41594-024-01224-7
- Primary Citation of Related Structures:
8IEB, 8IEC, 8IED, 8IEI, 8IEP, 8IEQ - PubMed Abstract:
Class C G-protein-coupled receptors (GPCRs) are activated through binding of agonists to the large extracellular domain (ECD) followed by rearrangement of the transmembrane domains (TMDs). GPR156, a class C orphan GPCR, is unique because it lacks an ECD and exhibits constitutive activity. Impaired GPR156-G i signaling contributes to loss of hearing. Here we present the cryo-electron microscopy structures of human GPR156 in the G o -free and G o -coupled states. We found that an endogenous phospholipid molecule is located within each TMD of the GPR156 dimer. Asymmetric binding of Gα to the phospholipid-bound GPR156 dimer restructures the first and second intracellular loops and the carboxy-terminal part of the elongated transmembrane 7 (TM7) without altering dimer conformation. Our findings reveal that GPR156 is a transducer for phospholipid signaling. Constant binding of abundant phospholipid molecules and the G-protein-induced reshaping of the cytoplasmic face provide a basis for the constitutive activation of GPR156.
- Department of Life Sciences, Pohang University of Science and Technology, Pohang, Republic of Korea.
Organizational Affiliation: