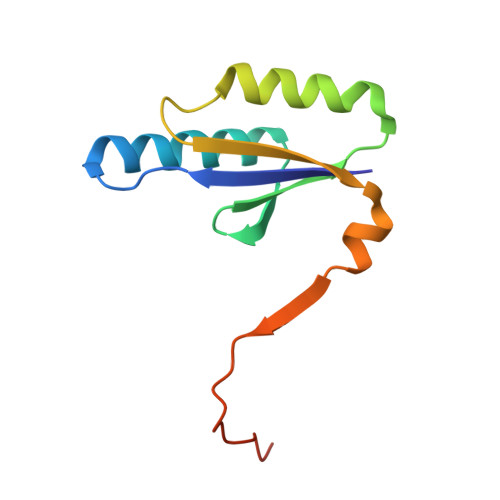
Insights into the inhibition of protospacer integration via direct interaction between Cas2 and AcrVA5.
Bi, M., Su, W., Li, J., Mo, X.(2024) Nat Commun 15: 3256-3256
- PubMed: 38627399
- DOI: https://doi.org/10.1038/s41467-024-47713-7
- Primary Citation of Related Structures:
8IA4 - PubMed Abstract:
Spacer acquisition step in CRISPR-Cas system involves the recognition and subsequent integration of protospacer by the Cas1-Cas2 complex in CRISPR-Cas systems. Here we report an anti-CRISPR protein, AcrVA5, and reveal the mechanisms by which it strongly inhibits protospacer integration. Our biochemical data shows that the integration by Cas1-Cas2 was abrogated in the presence of AcrVA5. AcrVA5 exhibits low binding affinity towards Cas2 and acetylates Cas2 at Lys 55 on the binding interface of the Cas2 and AcrVA5 N-terminal peptide complex to inhibit the Cas2-mediated endonuclease activity. Moreover, a detailed structural comparison between our crystal structure and homolog structure shows that binding of AcrVA5 to Cas2 causes steric hindrance to the neighboring protospacer resulting in the partial disassembly of the Cas1-Cas2 and protospacer complex, as demonstrated by electrophoretic mobility shift assay. Our study focuses on this mechanism of spacer acquisition inhibition and provides insights into the biology of CRISPR-Cas systems.
- College of Veterinary Medicine, Jilin University, 130062, Changchun, Jilin, China.
Organizational Affiliation: