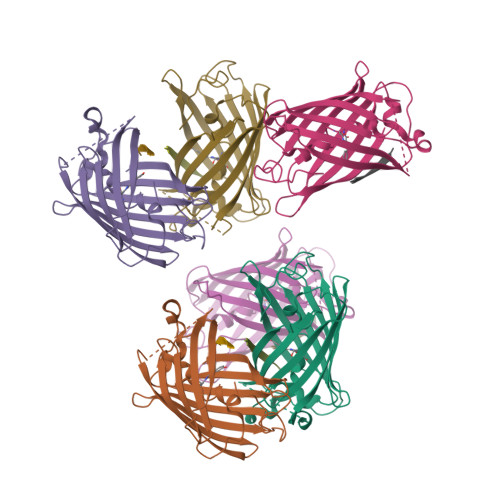
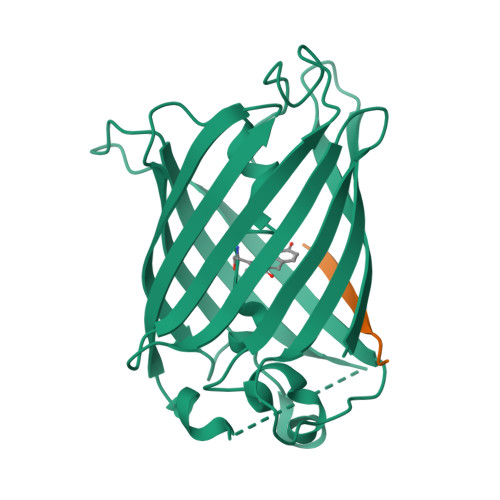
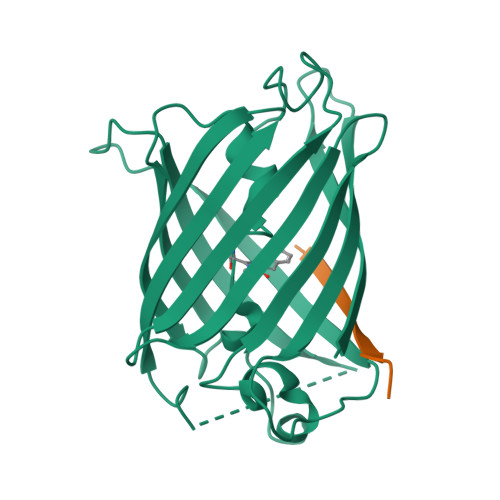
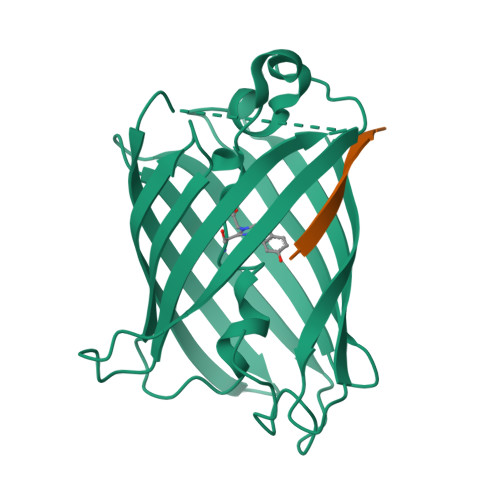
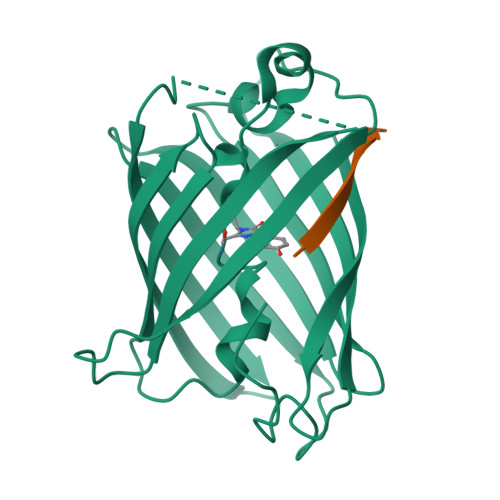
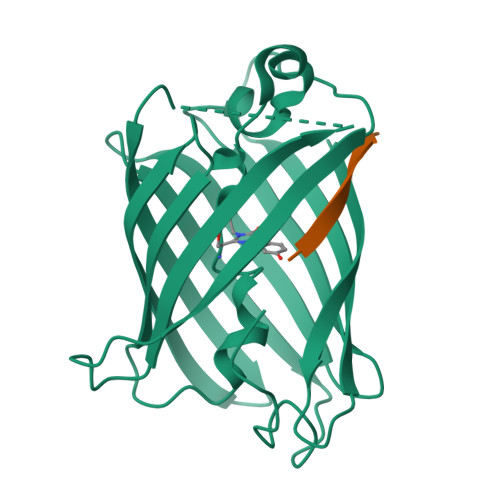
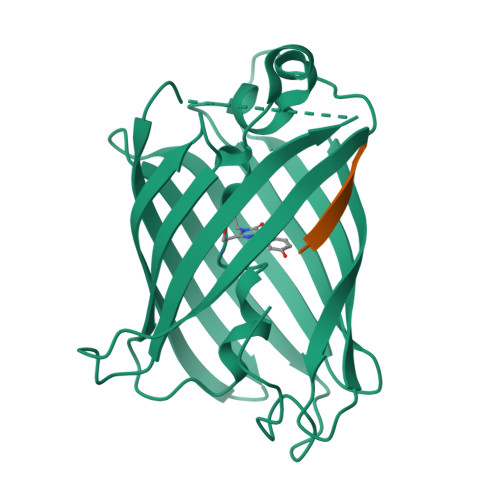
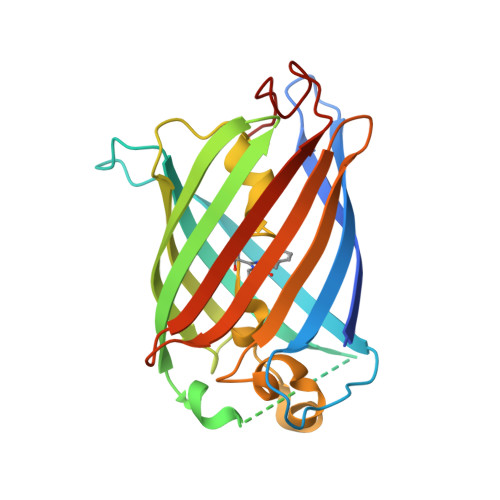
Engineering of a Fluorescent Protein for a Sensing of an Intrinsically Disordered Protein through Transition in the Chromophore State.
Yu, T.G., Lee, J., Yoon, J., Choi, J.M., Kim, D.G., Heo, W.D., Song, J.J., Kim, H.S.(2023) JACS Au 3: 3055-3065
- PubMed: 38034956
- DOI: https://doi.org/10.1021/jacsau.3c00445
- Primary Citation of Related Structures:
8I4O - PubMed Abstract:
Intrinsically disordered proteins (IDPs) not only play important roles in biological processes but are also linked with the pathogenesis of various human diseases. Specific and reliable sensing of IDPs is crucial for exploring their roles but remains elusive due to structural plasticity. Here, we present the development of a new type of fluorescent protein for the ratiometric sensing and tracking of an IDP. A β-strand of green fluorescent protein (GFP) was truncated, and the resulting GFP was further engineered to undergo the transition in the absorption maximum upon binding of a target motif within amyloid-β (Aβ) as a model IDP through rational design and directed evolution. Spectroscopic and structural analyses of the engineered truncated GFP demonstrated that a shift in the absorption maximum is driven by the change in the chromophore state from an anionic (460 nm) state into a neutral (390 nm) state as the Aβ binds, allowing a ratiometric detection of Aβ. The utility of the developed GFP was shown by the efficient and specific detection of an Aβ and the tracking of its conformational change and localization in astrocytes.
Organizational Affiliation:
Departement of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), Daejeon 34141, Korea.