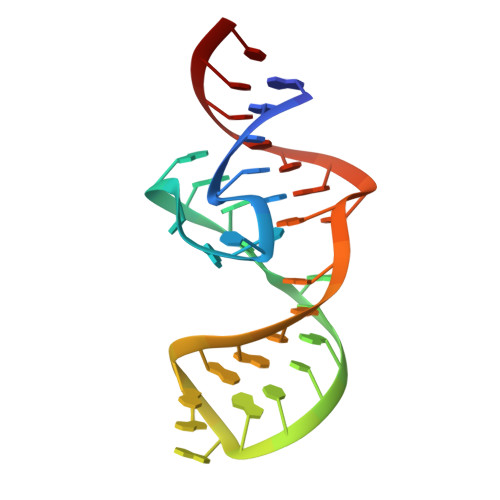
Structural basis of a small monomeric Clivia fluorogenic RNA with a large Stokes shift.
Huang, K., Song, Q., Fang, M., Yao, D., Shen, X., Xu, X., Chen, X., Zhu, L., Yang, Y., Ren, A.(2024) Nat Chem Biol 20: 1453-1460
- PubMed: 38816645
- DOI: https://doi.org/10.1038/s41589-024-01633-1
- Primary Citation of Related Structures:
8HZD, 8HZE, 8HZF, 8HZJ, 8HZK, 8HZL, 8HZM - PubMed Abstract:
RNA-based fluorogenic modules have revolutionized the spatiotemporal localization of RNA molecules. Recently, a fluorophore named 5-((Z)-4-((2-hydroxyethyl)(methyl)amino)benzylidene)-3-methyl-2-((E)-styryl)-3,5-dihydro-4H-imidazol-4-one (NBSI), emitting in red spectrum, and its cognate aptamer named Clivia were identified, exhibiting a large Stokes shift. To explore the underlying molecular basis of this unique RNA-fluorophore complex, we determined the tertiary structure of Clivia-NBSI. The overall structure uses a monomeric, non-G-quadruplex compact coaxial architecture, with NBSI sandwiched at the core junction. Structure-based fluorophore recognition pattern analysis, combined with fluorescence assays, enables the orthogonal use of Clivia-NBSI and other fluorogenic aptamers, paving the way for both dual-emission fluorescence and bioluminescence imaging of RNA molecules within living cells. Furthermore, on the basis of the structure-based substitution assay, we developed a multivalent Clivia fluorogenic aptamer containing multiple minimal NBSI-binding modules. This innovative design notably enhances the recognition sensitivity of fluorophores both in vitro and in vivo, shedding light on future efficient applications in various biomedical and research contexts.
- Department of Cardiology, The Second Affiliated Hospital of School of Medicine, Zhejiang University, Hangzhou, China.
Organizational Affiliation: