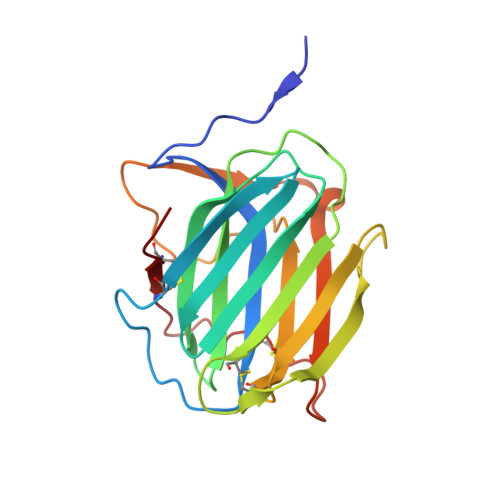
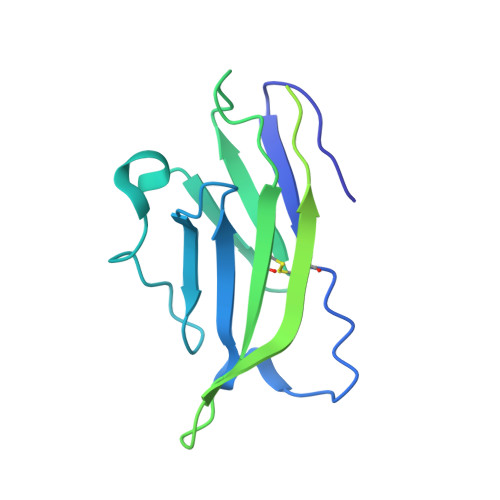
Structural and functional insights into the modulation of T cell costimulation by monkeypox virus protein M2.
Yang, S., Wang, Y., Yu, F., Cheng, R., Zhang, Y., Zhou, D., Ren, X., Deng, Z., Zhao, H.(2023) Nat Commun 14: 5186-5186
- PubMed: 37626059
- DOI: https://doi.org/10.1038/s41467-023-40748-2
- Primary Citation of Related Structures:
8HXA, 8HXB, 8HXC - PubMed Abstract:
The rapid spread of monkeypox in multiple countries has resulted in a global public health threat and has caused international concerns since May 2022. Poxvirus encoded M2 protein is a member of the poxvirus immune evasion family and plays roles in host immunomodulation via the regulation of innate immune response mediated by the NF-κB pathway and adaptive immune response mediated by B7 ligands. However, the interaction of monkeypox virus (MPXV) M2 with B7 ligands and structural insight into poxviral M2 function have remained elusive. Here we reveal that MPXV M2, co-existing as a hexamer and a heptamer, recognizes human B7.1 and B7.2 (hB7.1/2) with high avidities. The binding of oligomeric MPXV M2 interrupts the interactions of hB7.1/2 with CD28 and CTLA4 and subverts T cell activation mediated by B7.1/2 costimulatory signals. Cryo-EM structures of M2 in complex with hB7.1/2 show that M2 binds to the shallow concave face of hB7.1/2 and displays sterically competition with CD28 and CTLA4 for the binding to hB7.1/2. Our findings provide structural mechanisms of poxviral M2 function and immune evasion deployed by poxviruses.
- State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan, Hubei, China.
Organizational Affiliation: