Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Fang, J., Chai, Z., Huang, R., Huang, C., Ming, Z., Chen, B., Yao, W., Zhang, M.(2023) Front Plant Sci 14: 1191449-1191449
- PubMed: 37304725
- DOI: https://doi.org/10.3389/fpls.2023.1191449
- Primary Citation of Related Structures:
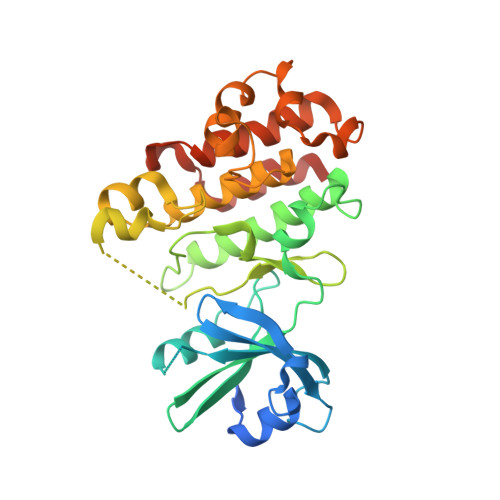
8HO6, 8HOA, 8HOD - PubMed Abstract:
Receptor-like cytoplastic kinases (RLCKs) are known in many plants to be involved in various processes of plant growth and development and regulate plant immunity to pathogen infection. Environmental stimuli such as pathogen infection and drought restrict the crop yield and interfere with plant growth. However, the function of RLCKs in sugarcane remains unclear. In this study, a member of the RLCK VII subfamily, ScRIPK, was identified in sugarcane based on sequence similarity to the rice and Arabidopsis RLCKs. ScRIPK was localized to the plasma membrane, as predicted, and the expression of ScRIPK was responsive to polyethylene glycol treatment and Fusarium sacchari infection. Overexpression of ScRIPK in Arabidopsis enhanced drought tolerance and disease susceptibility of seedlings. Moreover, the crystal structure of the ScRIPK kinase domain (ScRIPK KD) and the mutant proteins (ScRIPK-KD K124R and ScRIPK-KD S253A|T254A) were characterized in order to determine the activation mechanism. We also identified ScRIN4 as the interacting protein of ScRIPK. Our work identified a RLCK in sugarcane, providing a potential target for sugarcane responses to disease infection and drought, and a structural basis for kinase activation mechanisms.
- College of Agricultural, Guangxi University, Nanning, China.
Organizational Affiliation: