Structure insights into Histamine H4 receptor activation by an endogenous ligand histamine and agonist imetit
Sun, X.Y., Li, F.H., Wang, J.Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(i) subunit alpha-1 | 354 | Homo sapiens | Mutation(s): 0 Gene Names: GNAI1 EC: 3.6.5 Membrane Entity: Yes | 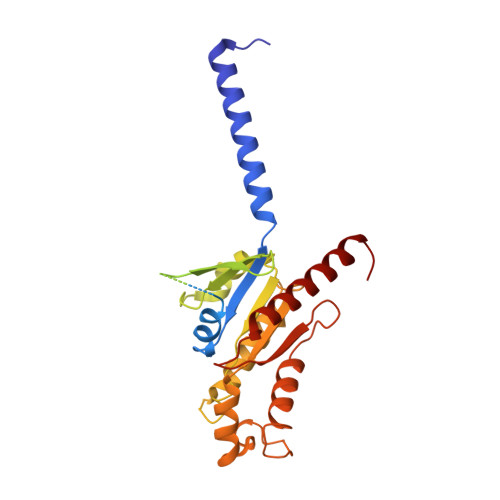 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P63096 (Homo sapiens) Explore P63096 Go to UniProtKB: P63096 | |||||
PHAROS: P63096 GTEx: ENSG00000127955 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63096 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | 340 | Homo sapiens | Mutation(s): 0 Gene Names: GNB1 | 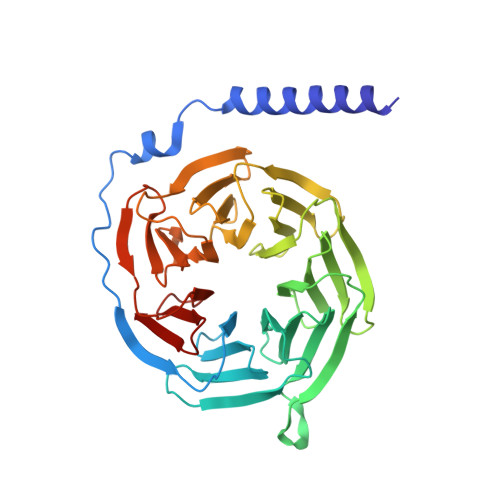 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P62873 (Homo sapiens) Explore P62873 Go to UniProtKB: P62873 | |||||
PHAROS: P62873 GTEx: ENSG00000078369 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62873 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 | C [auth G] | 71 | Homo sapiens | Mutation(s): 0 Gene Names: GNG2 Membrane Entity: Yes | 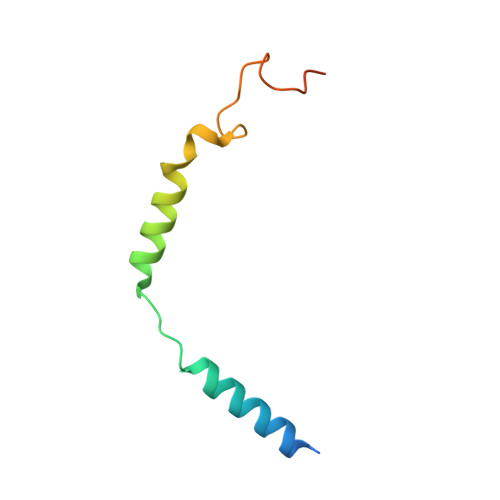 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P59768 (Homo sapiens) Explore P59768 Go to UniProtKB: P59768 | |||||
PHAROS: P59768 GTEx: ENSG00000186469 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P59768 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Soluble cytochrome b562, Histamine H4 receptor fusion | D [auth R] | 549 | Homo sapiens | Mutation(s): 0 Gene Names: cybC, HRH4, GPCR105 Membrane Entity: Yes | 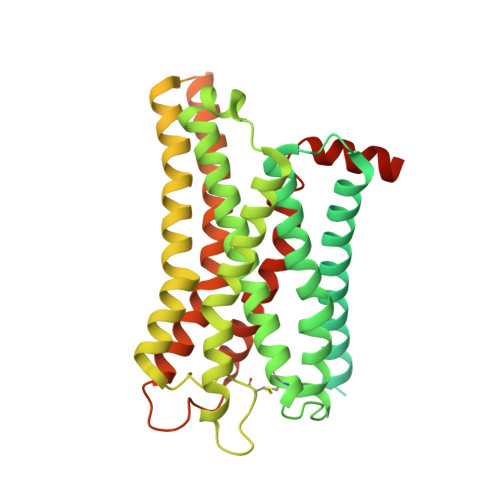 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P0ABE7 (Escherichia coli) Explore P0ABE7 Go to UniProtKB: P0ABE7 | |||||
Find proteins for Q9H3N8 (Homo sapiens) Explore Q9H3N8 Go to UniProtKB: Q9H3N8 | |||||
PHAROS: Q9H3N8 GTEx: ENSG00000134489 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | P0ABE7Q9H3N8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Single-chain variable fragment scFv16 | E [auth S] | 297 | Homo sapiens | Mutation(s): 0 |  |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HSM (Subject of Investigation/LOI) Query on HSM | F [auth R] | HISTAMINE C5 H9 N3 NTYJJOPFIAHURM-UHFFFAOYSA-N |  | ||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |