Structural Insight into the Mechanism of sigma 32-Mediated Transcription Initiation of Bacterial RNA Polymerase.
Lu, Q., Chen, T., Wang, J., Wang, F., Ye, W., Ma, L., Wu, S.(2023) Biomolecules 13
- PubMed: 37238608
- DOI: https://doi.org/10.3390/biom13050738
- Primary Citation of Related Structures:
8HKC - PubMed Abstract:
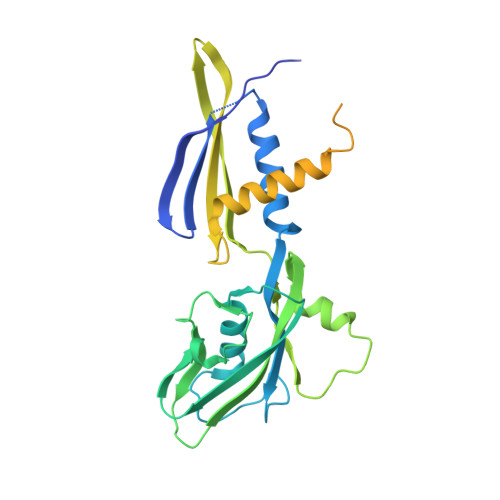
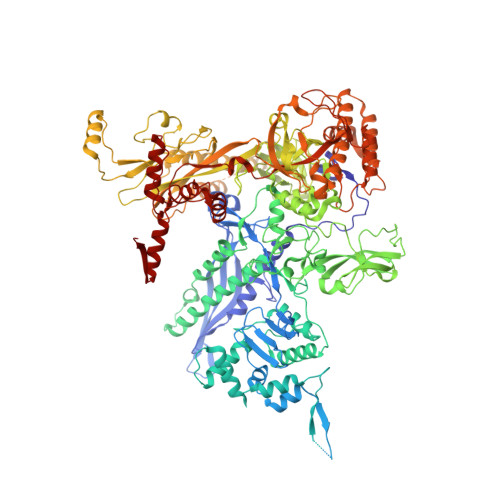
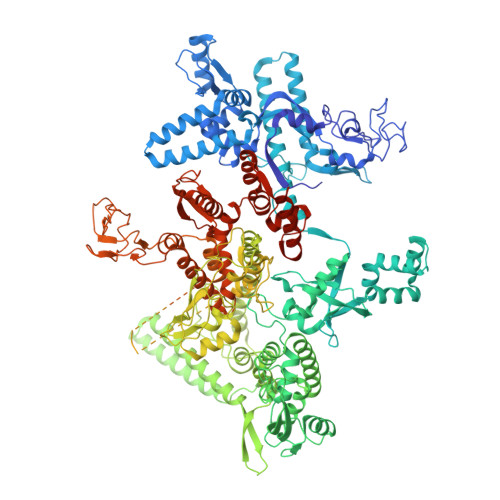
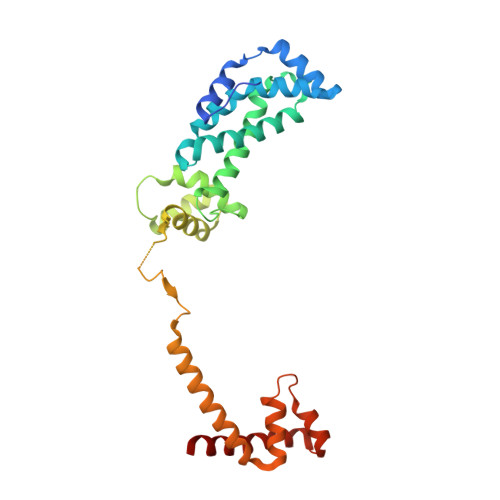
Bacterial RNA polymerases (RNAP) form distinct holoenzymes with different σ factors to initiate diverse gene expression programs. In this study, we report a cryo-EM structure at 2.49 Å of RNA polymerase transcription complex containing a temperature-sensitive bacterial σ factor, σ 32 (σ 32 -RPo). The structure of σ 32 -RPo reveals key interactions essential for the assembly of E. coli σ 32 -RNAP holoenzyme and for promoter recognition and unwinding by σ 32 . Specifically, a weak interaction between σ 32 and -35/-10 spacer is mediated by T128 and K130 in σ 32 . A histidine in σ 32 , rather than a tryptophan in σ 70 , acts as a wedge to separate the base pair at the upstream junction of the transcription bubble, highlighting the differential promoter-melting capability of different residue combinations. Structure superimposition revealed relatively different orientations between βFTH and σ 4 from other σ-engaged RNAPs and biochemical data suggest that a biased σ 4 -βFTH configuration may be adopted to modulate binding affinity to promoter so as to orchestrate the recognition and regulation of different promoters. Collectively, these unique structural features advance our understanding of the mechanism of transcription initiation mediated by different σ factors.
- State Key Laboratory of Biocatalysis and Enzyme Engineering, Hubei Collaborative Innovation Center for Green Transformation of Bio-Resources, Hubei Key Laboratory of Industrial Biotechnology, School of Life Sciences, Hubei University, Wuhan 430062, China.
Organizational Affiliation: