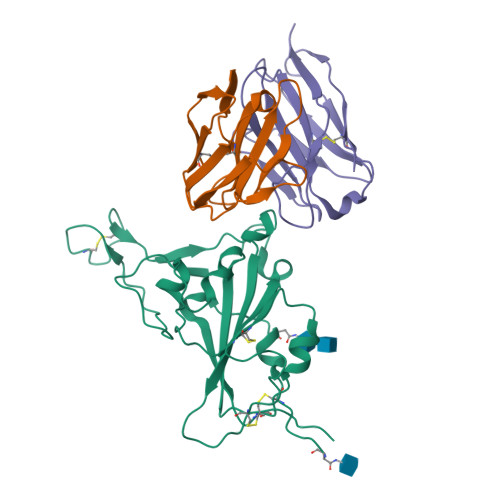
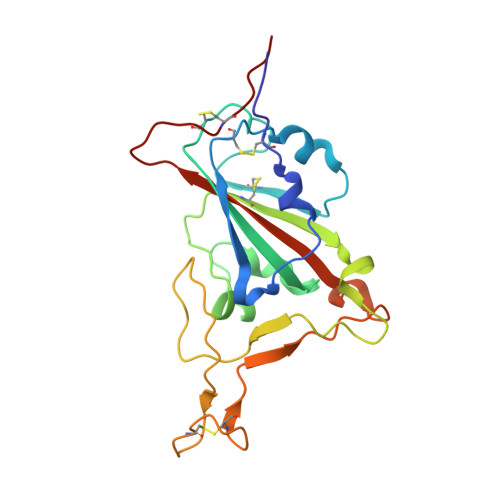
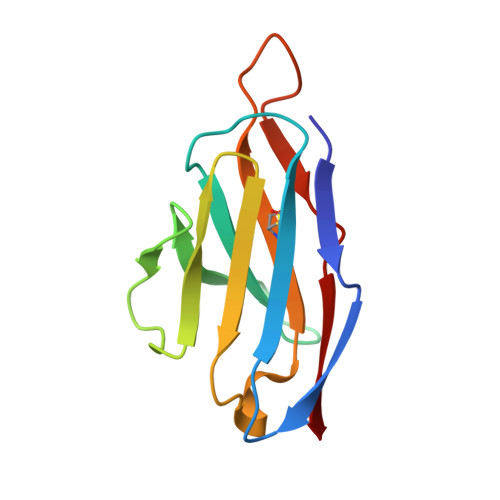
Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Chu, X., Ding, X., Yang, Y., Lu, Y., Li, T., Gao, Y., Zheng, L., Xiao, H., Yang, T., Cheng, H., Huang, H., Liu, Y., Lou, Y., Wu, C., Chen, Y., Yang, H., Ji, X., Guo, H.(2023) Biochem Biophys Res Commun 660: 43-49
- PubMed: 37062240
- DOI: https://doi.org/10.1016/j.bbrc.2023.04.002
- Primary Citation of Related Structures:
8HEB, 8HEC, 8HED - PubMed Abstract:
The COVID-19 pandemic, caused by SARS-CoV-2, has led to over 750 million infections and 6.8 million deaths worldwide since late 2019. Due to the continuous evolution of SARS-CoV-2, many significant variants have emerged, creating ongoing challenges to the prevention and treatment of the pandemic. Therefore, the study of antibody responses against SARS-CoV-2 is essential for the development of vaccines and therapeutics. Here we perform single particle cryo-electron microscopy (cryo-EM) structure determination of a rabbit monoclonal antibody (RmAb) 9H1 in complex with the SARS-CoV-2 wild-type (WT) spike trimer. Our structural analysis shows that 9H1 interacts with the receptor-binding motif (RBM) region of the receptor-binding domain (RBD) on the spike protein and by directly competing with angiotensin-converting enzyme 2 (ACE2), it blocks the binding of the virus to the receptor and achieves neutralization. Our findings suggest that utilizing rabbit-derived mAbs provides valuable insights into the molecular interactions between neutralizing antibodies and spike proteins and may also facilitate the development of therapeutic antibodies and expand the antibody library.
Organizational Affiliation:
The State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Nanjing University, Nanjing, Jiangsu, 210023, China.