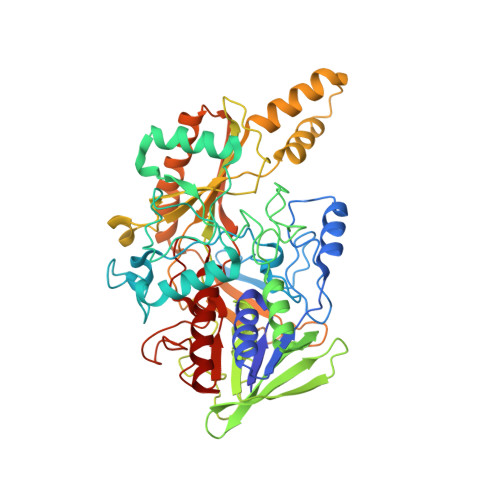
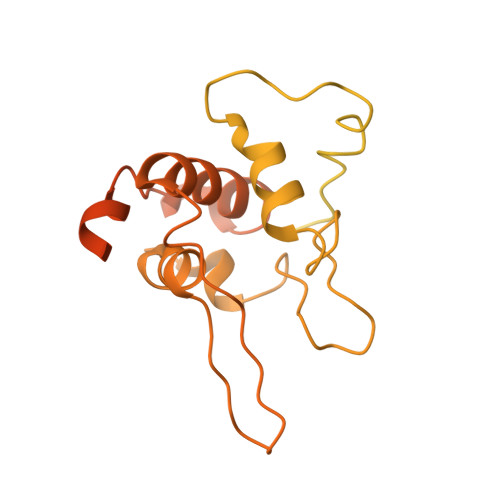
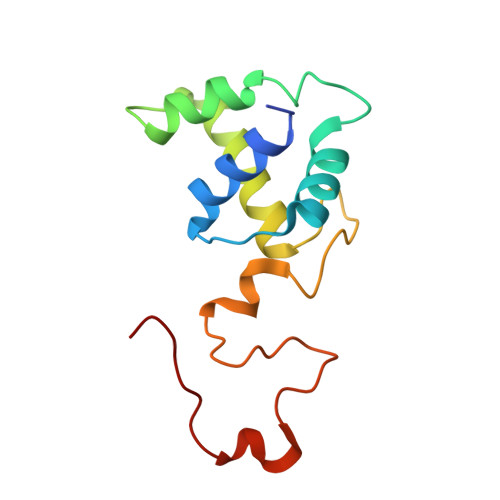
Microgravity environment grown crystal structure information based engineering of direct electron transfer type glucose dehydrogenase.
Okuda-Shimazaki, J., Yoshida, H., Lee, I., Kojima, K., Suzuki, N., Tsugawa, W., Yamada, M., Inaka, K., Tanaka, H., Sode, K.(2022) Commun Biol 5: 1334-1334
- PubMed: 36473944
- DOI: https://doi.org/10.1038/s42003-022-04286-9
- Primary Citation of Related Structures:
8HDD - PubMed Abstract:
The heterotrimeric flavin adenine dinucleotide dependent glucose dehydrogenase is a promising enzyme for direct electron transfer (DET) principle-based glucose sensors within continuous glucose monitoring systems. We elucidate the structure of the subunit interface of this enzyme by preparing heterotrimer complex protein crystals grown under a space microgravity environment. Based on the proposed structure, we introduce inter-subunit disulfide bonds between the small and electron transfer subunits (5 pairs), as well as the catalytic and the electron transfer subunits (9 pairs). Without compromising the enzyme's catalytic efficiency, a mutant enzyme harboring Pro205Cys in the catalytic subunit, Asp383Cys and Tyr349Cys in the electron transfer subunit, and Lys155Cys in the small subunit, is determined to be the most stable of the variants. The developed engineered enzyme demonstrate a higher catalytic activity and DET ability than the wild type. This mutant retains its full activity below 70 °C as well as after incubation at 75 °C for 15 min - much higher temperatures than the current gold standard enzyme, glucose oxidase, is capable of withstanding.
- Joint Department of Biomedical Engineering, The University of North Carolina at Chapel Hill and North Carolina State University, Chapel Hill, NC27599, USA.
Organizational Affiliation: