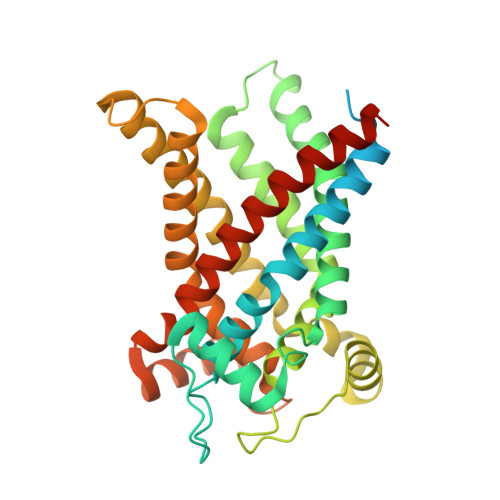
Structural basis for the binding of DNP and purine nucleotides onto UCP1.
Kang, Y., Chen, L.(2023) Nature 620: 226-231
- PubMed: 37336486
- DOI: https://doi.org/10.1038/s41586-023-06332-w
- Primary Citation of Related Structures:
8HBV, 8HBW, 8J1N - PubMed Abstract:
Uncoupling protein 1 (UCP1) conducts protons through the inner mitochondrial membrane to uncouple mitochondrial respiration from ATP production, thereby converting the electrochemical gradient of protons into heat 1,2 . The activity of UCP1 is activated by endogenous fatty acids and synthetic small molecules, such as 2,4-dinitrophenol (DNP), and is inhibited by purine nucleotides, such as ATP 3-5 . However, the mechanism by which UCP1 binds to these ligands remains unknown. Here we present the structures of human UCP1 in the nucleotide-free state, the DNP-bound state and the ATP-bound state. The structures show that the central cavity of UCP1 is open to the cytosolic side. DNP binds inside the cavity, making contact with transmembrane helix 2 (TM2) and TM6. ATP binds in the same cavity and induces conformational changes in TM2, together with the inward bending of TM1, TM4, TM5 and TM6 of UCP1, resulting in a more compact structure of UCP1. The binding site of ATP overlaps with that of DNP, suggesting that ATP competitively blocks the functional engagement of DNP, resulting in the inhibition of the proton-conducting activity of UCP1.
- State Key Laboratory of Membrane Biology, College of Future Technology, Institute of Molecular Medicine, Peking University, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Beijing, China.
Organizational Affiliation: