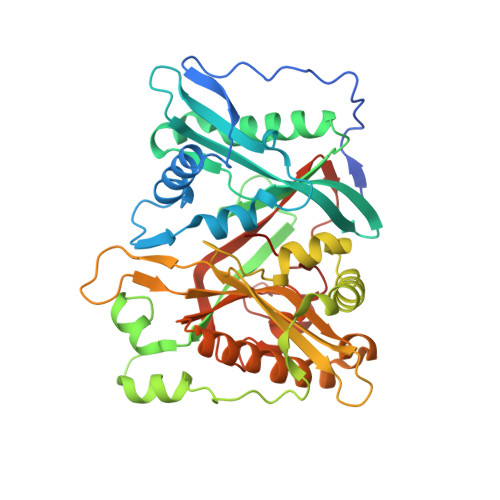
N -Myristoyltransferase, a Potential Antifungal Candidate Drug-Target for Aspergillus flavus.
Wang, Y., Lin, R., Liu, M., Wang, S., Chen, H., Zeng, W., Nie, X., Wang, S.(2023) Microbiol Spectr 11: e0421222-e0421222
- PubMed: 36541770
- DOI: https://doi.org/10.1128/spectrum.04212-22
- Primary Citation of Related Structures:
8HBS - PubMed Abstract:
The filamentous fungus Aspergillus flavus causes devastating diseases not only to cash crops but also to humans by secreting a series of secondary metabolites called aflatoxins. In the cotranslational or posttranslational process, N -myristoyltransferase (Nmt) is a crucial enzyme that catalyzes the myristate group from myristoyl-coenzyme A (myristoyl-CoA) to the N terminus or internal glycine residue of a protein by forming a covalent bond. Members of the Nmt family execute a diverse range of biological functions across a broad range of fungi. However, the underlying mechanism of AflNmt action in the A. flavus life cycle is unclear, particularly during the growth, development, and secondary metabolic synthesis stages. In the present study, AlfNmt was found to be essential for the development of spore and sclerotia, based on the regulation of the xylose-inducible promoter. AflNmt, located in the cytoplasm of A. flavus, is also involved in modulating aflatoxin (AFB1) in A. flavus, which has not previously been reported in Aspergillus spp. In addition, we purified, characterized, and crystallized the recombinant AflNmt protein (rAflNmt) from the Escherichia coli expression system. Interestingly, the crystal structure of rAlfNmt is moderately different from the models predicted by AlphaFold2 in the N-terminal region, indicating the limitations of machine-learning prediction. In conclusion, these results provide a molecular basis for the functional role of AflNmt in A. flavus and structural insights concerning protein prediction. IMPORTANCE As an opportunistic pathogen, A. flavus causes crop loss due to fungal growth and mycotoxin contamination. Investigating the role of virulence factors during infection and searching for novel drug targets have been popular scientific topics in the field of fungal control. Nmt has become a potential target in some organisms. However, whether Nmt is involved in the developmental stages of A. flavus and aflatoxin synthesis, and whether AlfNmt is an ideal target for structure-based drug design, remains unclear. This study systematically explored and identified the role of AlfNmt in the development of spore and sclerotia, especially in aflatoxin biosynthesis. Moreover, although there is not much difference between the AflNmt model predicted using the AlphaFold2 technique and the structure determined using the X-ray method, current AI prediction models may not be suitable for structure-based drug development. There is still room for further improvements in protein prediction.
- State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China.
Organizational Affiliation: