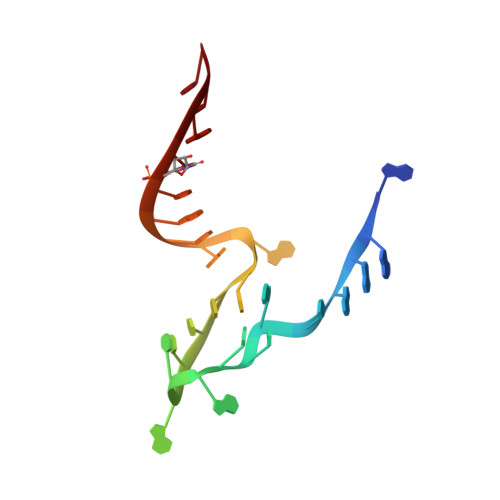
Crystal structures of the NAD+-II riboswitch reveal two distinct ligand-binding pockets.
Peng, X., Liao, W., Lin, X., Lilley, D.M.J., Huang, L.(2023) Nucleic Acids Res 51: 2904-2914
- PubMed: 36840714
- DOI: https://doi.org/10.1093/nar/gkad102
- Primary Citation of Related Structures:
8HB1, 8HB3, 8HB8, 8HBA, 8I3Z - PubMed Abstract:
We present crystal structures of a new NAD+-binding riboswitch termed NAD+-II, bound to nicotinamide mononucleotide (NMN), nicotinamide adenine dinucleotide (NAD+) and nicotinamide riboside (NR). The RNA structure comprises a number of structural features including three helices, one of which forms a triple helix by interacting with an A5 strand in its minor-groove, and another formed from a long-range pseudoknot. The core of the structure (centrally located and coaxial with the triplex and the pseudoknot) includes two consecutive quadruple base interactions. Unusually the riboswitch binds two molecules of ligand, bound at distinct, non-overlapping sites in the RNA. Binding occurs primarily through the nicotinamide moiety of each ligand, held by specific hydrogen bonding and stacking interactions with the pyridyl ring. The mode of binding is the same for NMN, NR and the nicotinamide moiety of NAD+. In addition, when NAD+ is bound into one site it adopts an elongated conformation such that its diphosphate linker occupies a groove on the surface of the RNA, following which the adenine portion inserts into a pocket and makes specific hydrogen bonding interactions. Thus the NAD+-II riboswitch is distinct from the NAD+-I riboswitch in that it binds two molecules of ligand at separate sites, and that binding occurs principally through the nicotinamide moiety.
- Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Guangdong-Hong Kong Joint Laboratory for RNA Medicine, Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University, Guangzhou, China.
Organizational Affiliation: